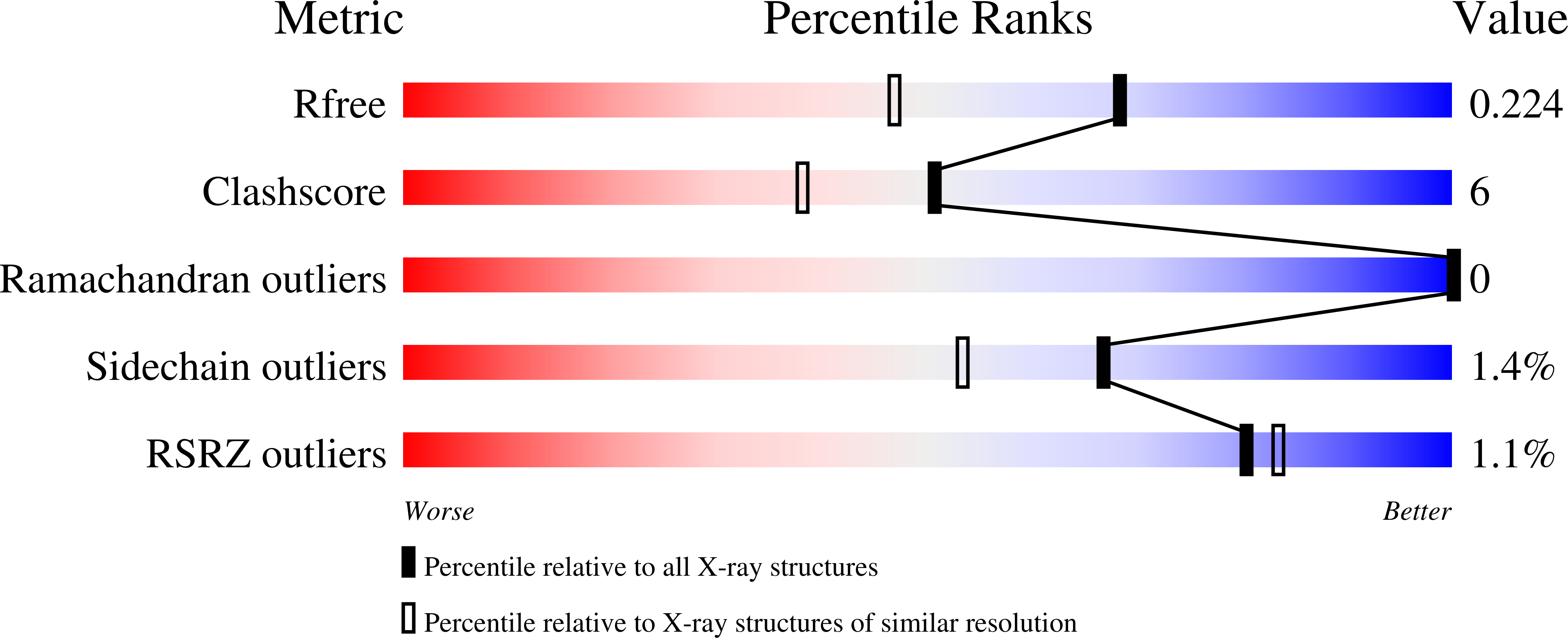
Crystal structure of the Fusarium oxysporum tannase-like feruloyl esterase FaeC in complex with p-coumaric acid provides insight into ligand binding.
Ferousi, C., Kosinas, C., Nikolaivits, E., Topakas, E., Dimarogona, M.(2023) FEBS Lett 597: 1415-1427
- PubMed: 36961270
- DOI: https://doi.org/10.1002/1873-3468.14615
- Primary Citation of Related Structures:
8BHH - PubMed Abstract:
Feruloyl esterases (FAEs) hydrolyze the ester bonds between hydroxycinnamic acids and arabinose residues of plant cell walls and exhibit considerable diversity in terms of substrate specificity. Here, we report the crystal structure of an FAE from Fusarium oxysporum (FoFaeC) at 1.7 Å resolution in complex with p-coumaric acid, which is the first ligand-bound structure of a tannase-like FAE. Our data reveal local conformational changes around the active site upon ligand binding, suggesting alternation between an active and a resting state of the enzyme. A swinging tyrosine residue appears to be gating the substrate binding pocket, while the lid domain of the protein exerts substrate specificity by means of a well-defined hydrophobic core that encases the phenyl moiety of the substrate.
Organizational Affiliation:
Industrial Biotechnology & Biocatalysis Group, Biotechnology Laboratory, School of Chemical Engineering, National Technical University of Athens, Greece.