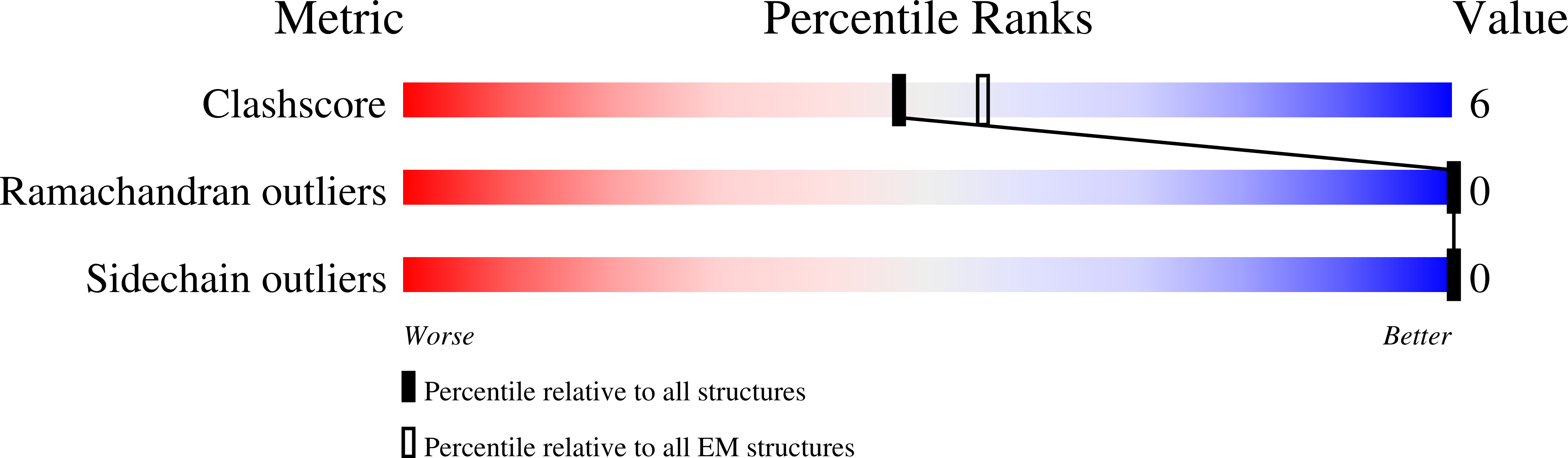Modulatory mechanisms of TARP gamma 8-selective AMPA receptor therapeutics.
Zhang, D., Lape, R., Shaikh, S.A., Kohegyi, B.K., Watson, J.F., Cais, O., Nakagawa, T., Greger, I.H.(2023) Nat Commun 14: 1659-1659
- PubMed: 36966141
- DOI: https://doi.org/10.1038/s41467-023-37259-5
- Primary Citation of Related Structures:
8AYL, 8AYM, 8AYN, 8AYO - PubMed Abstract:
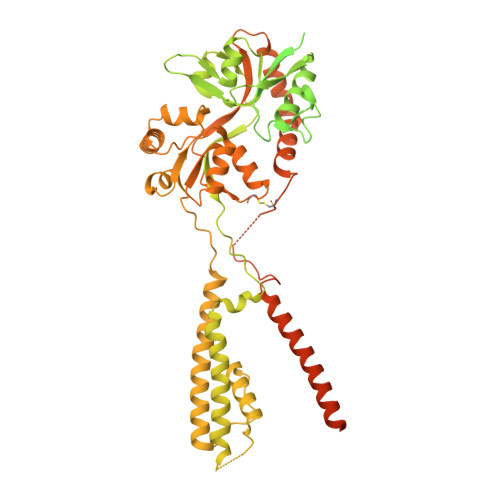
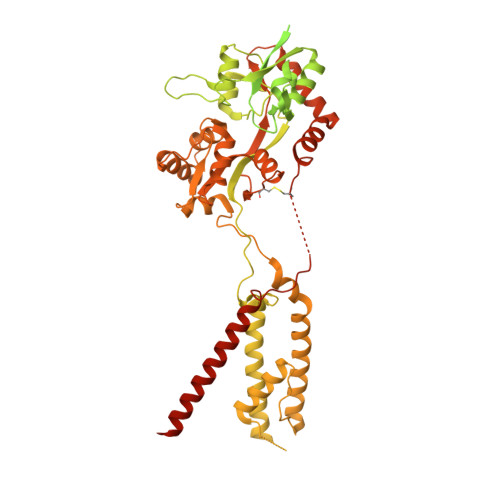
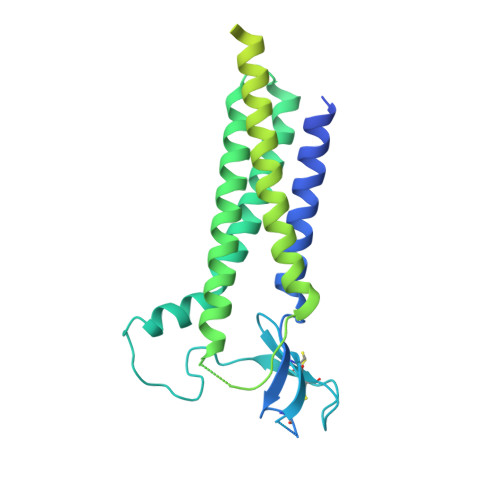
AMPA glutamate receptors (AMPARs) mediate excitatory neurotransmission throughout the brain. Their signalling is uniquely diversified by brain region-specific auxiliary subunits, providing an opportunity for the development of selective therapeutics. AMPARs associated with TARP γ8 are enriched in the hippocampus, and are targets of emerging anti-epileptic drugs. To understand their therapeutic activity, we determined cryo-EM structures of the GluA1/2-γ8 receptor associated with three potent, chemically diverse ligands. We find that despite sharing a lipid-exposed and water-accessible binding pocket, drug action is differentially affected by binding-site mutants. Together with patch-clamp recordings and MD simulations we also demonstrate that ligand-triggered reorganisation of the AMPAR-TARP interface contributes to modulation. Unexpectedly, one ligand (JNJ-61432059) acts bifunctionally, negatively affecting GluA1 but exerting positive modulatory action on GluA2-containing AMPARs, in a TARP stoichiometry-dependent manner. These results further illuminate the action of TARPs, demonstrate the sensitive balance between positive and negative modulatory action, and provide a mechanistic platform for development of both positive and negative selective AMPAR modulators.
Organizational Affiliation:
Neurobiology Division, MRC Laboratory of Molecular Biology, Cambridge, UK.