Crystal structure of PpSB1-LOV-K117E mutant (light state)
Batra-Safferling, R., Granzin, J., Krauss, U.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Sensory box protein | 162 | Pseudomonas putida KT2440 | Mutation(s): 1 Gene Names: PP_4629 | 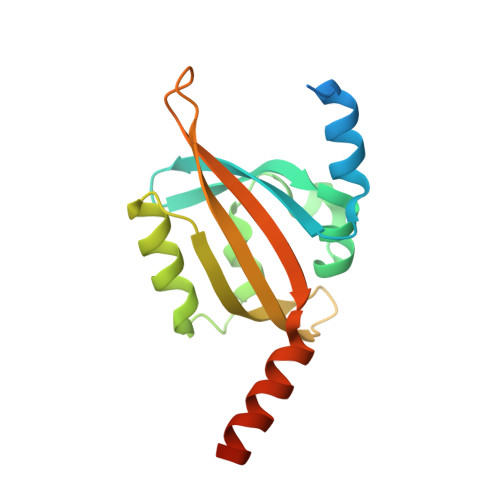 | |
UniProt | |||||
Find proteins for Q88E39 (Pseudomonas putida (strain ATCC 47054 / DSM 6125 / CFBP 8728 / NCIMB 11950 / KT2440)) Explore Q88E39 Go to UniProtKB: Q88E39 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q88E39 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| JGC (Subject of Investigation/LOI) Query on JGC | B [auth A] | Flavin mononucleotide (semi-quinone intermediate) C17 H23 N4 O9 P JNFNEOXSUODPSY-DGAVXFQQSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 54.329 | α = 90 |
| b = 54.329 | β = 90 |
| c = 220.555 | γ = 120 |
| Software Name | Purpose |
|---|---|
| MxCuBE | data collection |
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| German Federal Ministry for Education and Research | Germany | FKZ 031A16 |