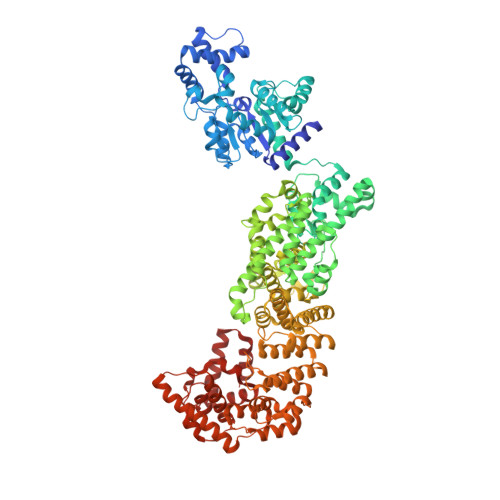
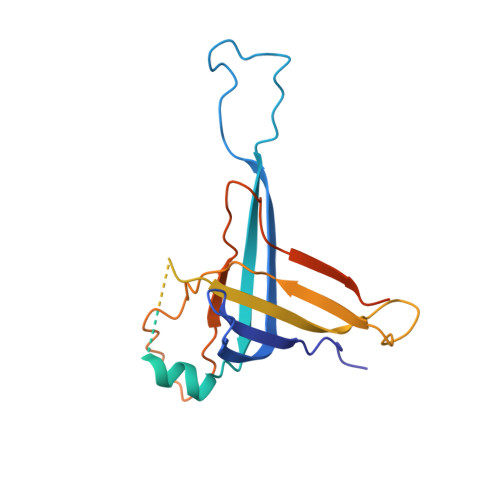
Structural insights into autoinhibition and activation of defense-associated sirtuin protein.
Yang, X., Wang, Y., Zheng, J.(2024) Int J Biol Macromol 277: 134145-134145
- PubMed: 39059542
- DOI: https://doi.org/10.1016/j.ijbiomac.2024.134145
- Primary Citation of Related Structures:
8YKF, 8YL5, 8YLN, 8YLT, 8Z18, 8ZTR - PubMed Abstract:
Bacterial defense-associated sirtuin 2 (DSR2) proteins harbor an N-terminal sirtuin (SIR2) domain degrading NAD + . DSR2 from Bacillus subtilis 29R is autoinhibited and unable to hydrolyze NAD + in the absence of phage infection. A tail tube protein (TTP) of phage SPR activates the DSR2 while a DSR2-inhibiting protein of phage SPbeta, known as DSAD1 (DSR anti-defense 1), inactivates the DSR2. Although DSR2 structures in complexed with TTP and DSAD1, respectively, have been reported recently, the autoinhibition and activation mechanisms remain incompletely understood. Here, we present cryo-electron microscopy structures of the DSR2-NAD + complex in autoinhibited state and the in vitro assembled DSR2-TFD (TTP tube-forming domain) complex in activated state. The DSR2-NAD + complex reveals that the autoinhibited DSR2 assembles into an inactive tetramer, binding NAD + through a distinct pocket situated outside active site. Binding of TFD into cavities within the sensor domains of DSR2 triggers a conformational change in SIR2 regions, activating its NADase activity, whereas the TTP β-sandwich domain (BSD) is flexible and does not contribute to the activation process. The activated form of DSR2 exists as tetramers and dimers, with the tetramers exhibiting more NADase activity. Overall, our results extend the current understanding of autoinhibition and activation of DSR2 immune proteins.
Organizational Affiliation:
State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 200240, China.