Cryo-EM structure of DSR2-TTP
Zhang, H., Li, Z., Li, X.Z.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| SIR2-like domain-containing protein | 1,005 | Bacillus subtilis | Mutation(s): 0 Gene Names: B4122_2870 | 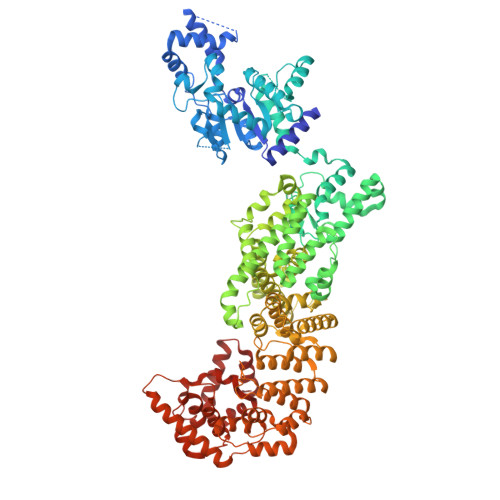 | |
UniProt | |||||
Find proteins for D4G637 (Bacillus subtilis subsp. natto (strain BEST195)) Go to UniProtKB: D4G637 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| tail tube protein(TTP) | 264 | Bacillus subtilis | Mutation(s): 0 Gene Names: B4122_1986 | 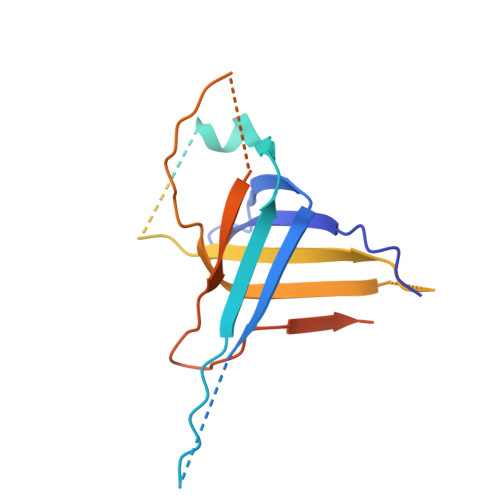 | |
UniProt | |||||
Find proteins for A0A162TY69 (Bacillus subtilis) Go to UniProtKB: A0A162TY69 | |||||
Sequence AnnotationsExpand | |||||
| |||||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Other government | -- |