A functionally augmented carbohydrate utilization locus from herbivore gut microbiota fueled by dietary beta-glucans.
Mandelli, F., Martins, M.P., Chinaglia, M., Lima, E.A., Morais, M.A.B., Lima, T.B., Cabral, L., Pirolla, R.A.S., Fuzita, F.J., Paixao, D.A.A., Andrade, M.O., Wolf, L.D., Vieira, P.S., Persinoti, G.F., Murakami, M.T.(2024) NPJ Biofilms Microbiomes 10: 105-105
- PubMed: 39397008
- DOI: https://doi.org/10.1038/s41522-024-00578-6
- Primary Citation of Related Structures:
8VA3, 8VA4, 8VA7 - PubMed Abstract:
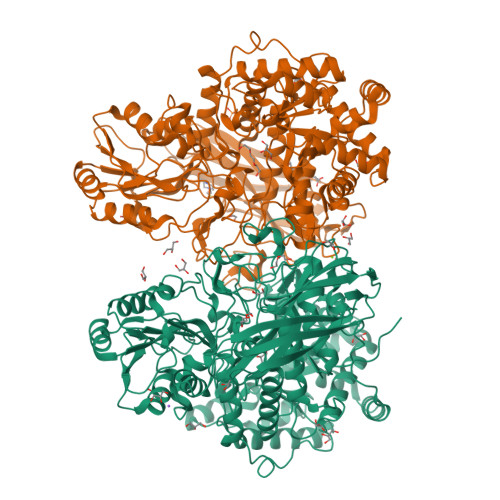
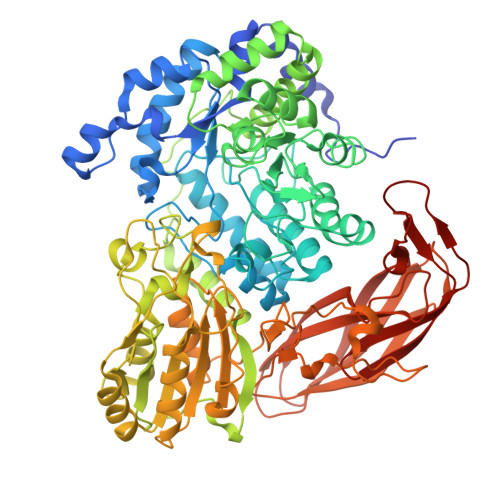
Gut microbiota members from the Bacteroidota phylum play a pivotal role in mammalian health and metabolism. They thrive in this diverse ecosystem due to their notable ability to cope with distinct recalcitrant dietary glycans via polysaccharide utilization loci (PULs). Our study reveals that a PUL from an herbivore gut bacterium belonging to the Bacteroidota phylum, with a gene composition similar to that in the human gut, exhibits extended functionality. While the human gut PUL targets mixed-linkage β-glucans specifically, the herbivore gut PUL also efficiently processes linear and substituted β-1,3-glucans. This gain of function emerges from molecular adaptations in recognition proteins and carbohydrate-active enzymes, including a β-glucosidase specialized for β(1,6)-glucosyl linkages, a typical substitution in β(1,3)-glucans. These findings broaden the existing model for non-cellulosic β-glucans utilization by gut bacteria, revealing an additional layer of functional and evolutionary complexity within the gut microbiota, beyond conventional gene insertions/deletions to intricate biochemical interactions.
Organizational Affiliation:
Brazilian Biorenewables National Laboratory (LNBR), Brazilian Center for Research in Energy and Materials (CNPEM), Campinas, São Paulo, 13083-970, Brazil.