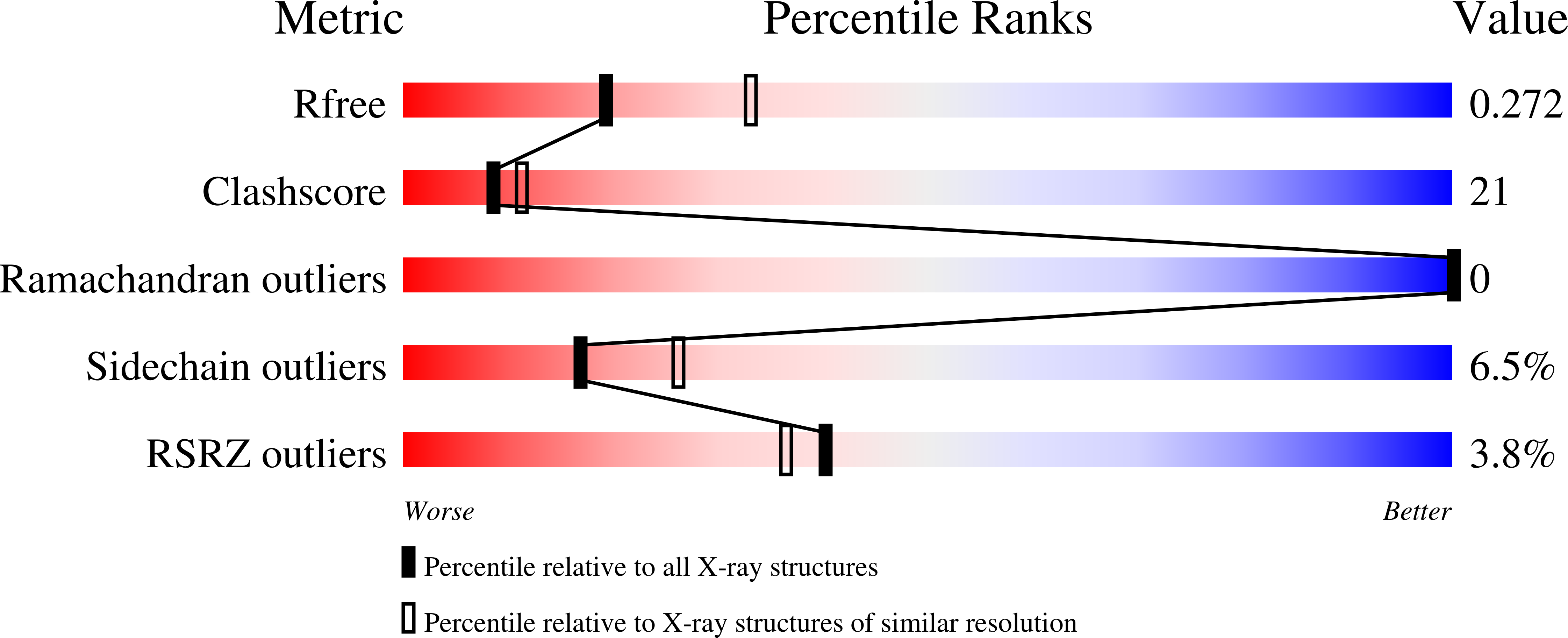
Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Prucha, G.R., Henry, S., Hollander, K., Carter, Z.J., Spasov, K.A., Jorgensen, W.L., Anderson, K.S.(2023) Eur J Med Chem 262: 115894-115894
- PubMed: 37883896
- DOI: https://doi.org/10.1016/j.ejmech.2023.115894
- Primary Citation of Related Structures:
8U69, 8U6A, 8U6B, 8U6C, 8U6D, 8U6E, 8U6F, 8U6G, 8U6H, 8U6I, 8U6J, 8U6K, 8U6L, 8U6M, 8U6N, 8U6O, 8U6P, 8U6Q, 8U6R, 8U6S, 8U6T - PubMed Abstract:
Reverse transcriptase (RT) is one of three key proteins responsible for the replication cycle of HIV-1 in the host. Several classes of inhibitors have been developed to target the enzyme, with non-nucleoside reverse transcriptase inhibitors forming first-line treatment. Previously, covalent RT inhibitors have been identified and found to bind irreversibly to commonly mutated residues such as Y181C. In this work we aim to circumvent the issue of NNRTI resistance through targeting K102, which has not yet been identified to confer drug resistance. As reported here, 34 compounds were synthesized and characterized biochemically and structurally with wild-type (WT) HIV-1 RT. Two of these inhibitors demonstrate covalent inhibition as evidenced by protein crystallography, enzyme kinetics, mass spectrometry, and antiviral potency in HIV-1 infected human T-cell assays.
Organizational Affiliation:
Department of Pharmacology, Yale University School of Medicine, New Haven, CT, 06520-8066, USA.