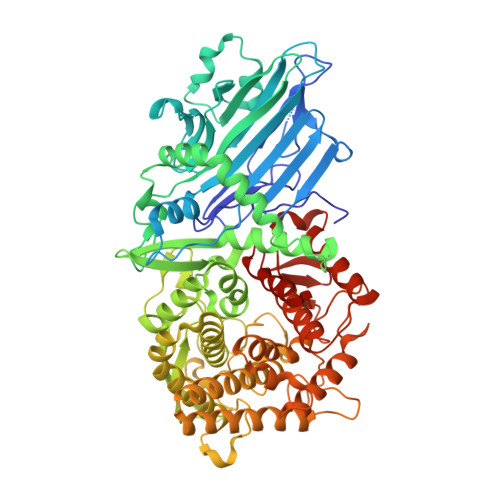
Structural Characterization of beta-Xylosidase XynB2 from Geobacillus stearothermophilus CECT43: A Member of the Glycoside Hydrolase Family GH52
Gavira, J.A., Contreras, L.M., Alshamaa, H.M., Clemente-Jimenez, J.M., Rodriguez-Vico, F., Las Heras-Vazquez, F.J., Martinez-Rodriguez, S.(2024) Crystals (Basel) 14