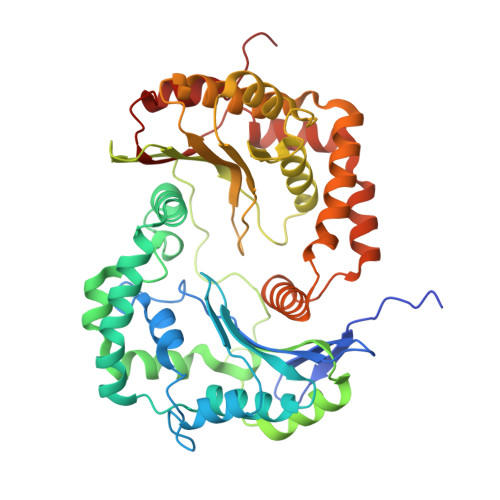
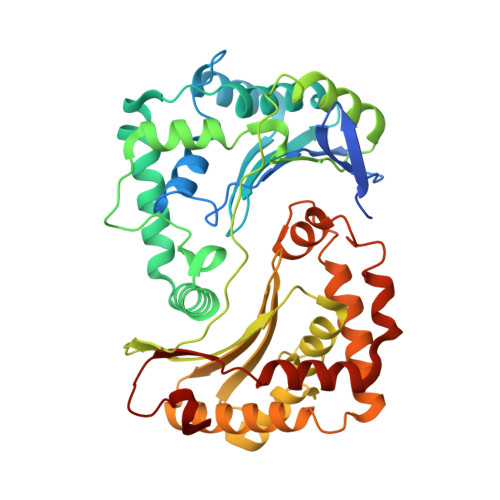
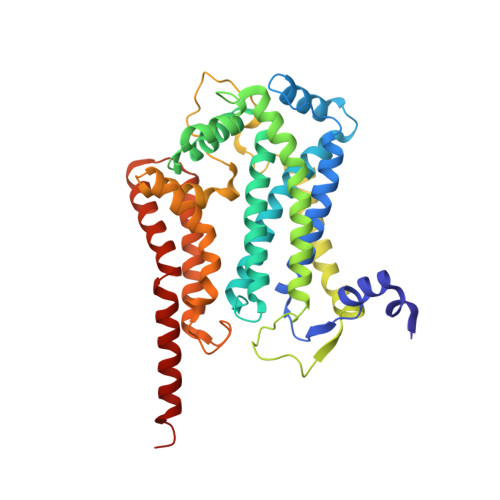
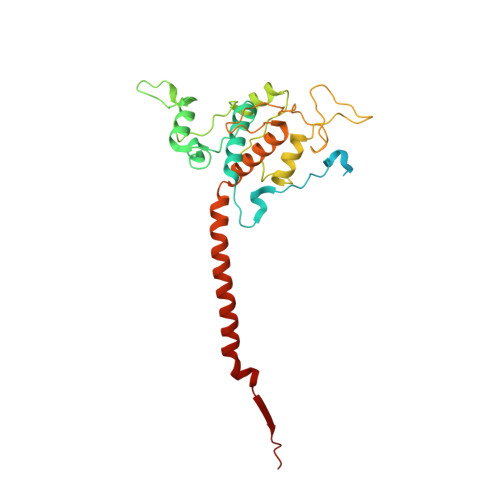
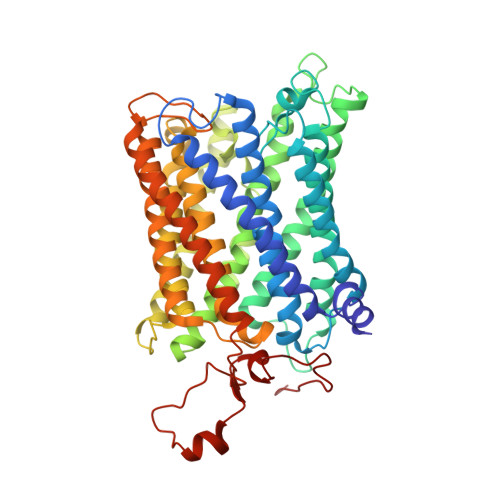
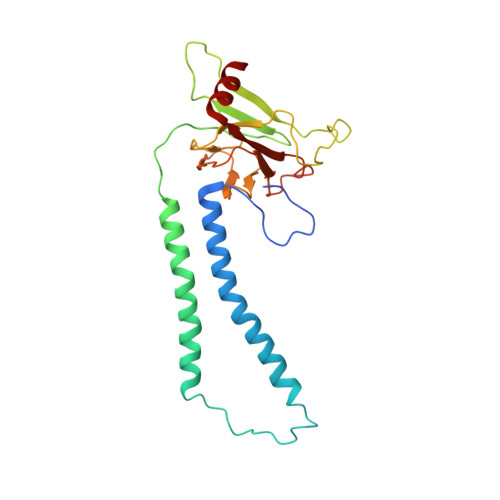
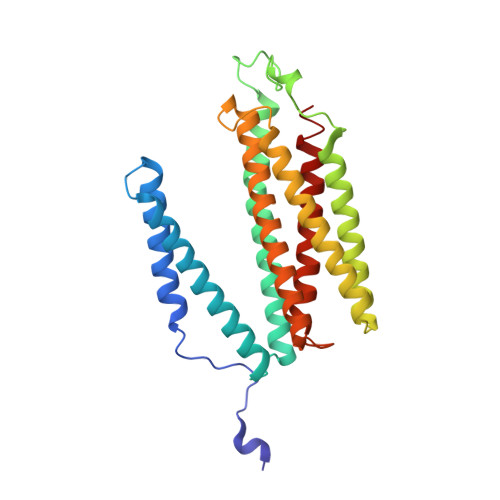
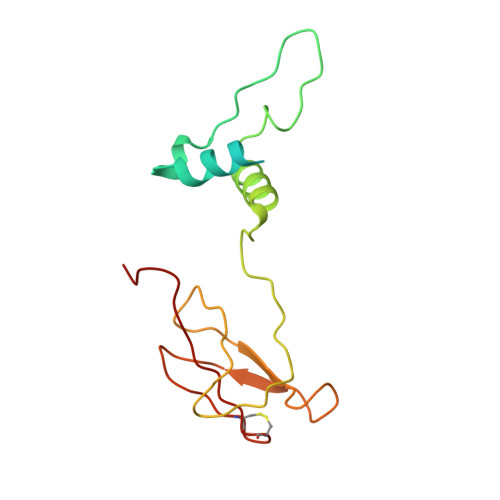
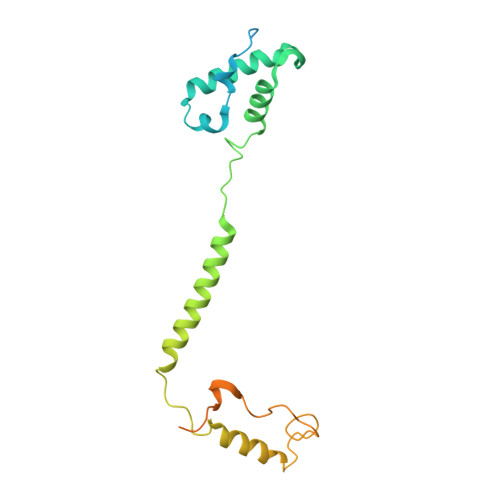
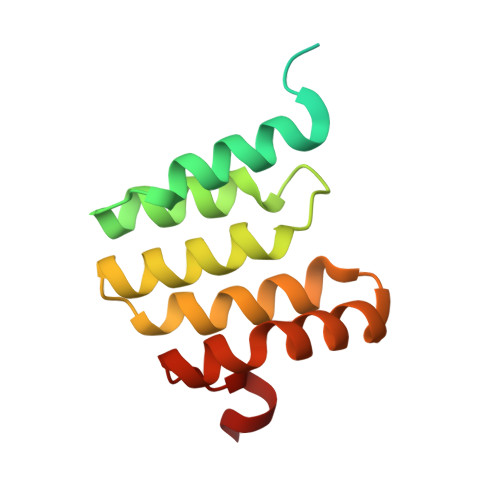
Structure and function of the S. pombe III-IV-cyt c supercomplex.
Moe, A., Dimogkioka, A.R., Rapaport, D., Ojemyr, L.N., Brzezinski, P.(2023) Proc Natl Acad Sci U S A 120: e2307697120-e2307697120
- PubMed: 37939086
- DOI: https://doi.org/10.1073/pnas.2307697120
- Primary Citation of Related Structures:
8Q1B - PubMed Abstract:
The respiratory chain in aerobic organisms is composed of a number of membrane-bound protein complexes that link electron transfer to proton translocation across the membrane. In mitochondria, the final electron acceptor, complex IV (CIV), receives electrons from dimeric complex III (CIII 2 ), via a mobile electron carrier, cytochrome c . In the present study, we isolated the CIII 2 CIV supercomplex from the fission yeast Schizosaccharomyces pombe and determined its structure with bound cyt. c using single-particle electron cryomicroscopy. A respiratory supercomplex factor 2 was found to be bound at CIV distally positioned in the supercomplex. In addition to the redox-active metal sites, we found a metal ion, presumably Zn 2+ , coordinated in the CIII subunit Cor1, which is encoded by the same gene ( qcr 1 ) as the mitochondrial-processing peptidase subunit β. Our data show that the isolated CIII 2 CIV supercomplex displays proteolytic activity suggesting a dual role of CIII 2 in S. pombe . As in the supercomplex from S. cerevisiae , subunit Cox5 of CIV faces towards one CIII monomer, but in S. pombe, the two complexes are rotated relative to each other by ~45°. This orientation yields equal distances between the cyt. c binding sites at CIV and at each of the two CIII monomers. The structure shows cyt. c bound at four positions, but only along one of the two symmetrical branches. Overall, this combined structural and functional study reveals the integration of peptidase activity with the CIII 2 respiratory system and indicates a two-dimensional cyt. c diffusion mechanism within the CIII 2 -CIV supercomplex.
Organizational Affiliation:
Department of Biochemistry and Biophysics, The Arrhenius Laboratories for Natural Sciences, Stockholm University, Stockholm SE-106 91, Sweden.