Crystal Structure of the Catalytic Domain of a Botulinum Neurotoxin Homologue from Enterococcus faecium : Potential Insights into Substrate Recognition.
Gregory, K.S., Hall, P.R., Onuh, J.P., Mojanaga, O.O., Liu, S.M., Acharya, K.R.(2023) Int J Mol Sci 24
- PubMed: 37628902
- DOI: https://doi.org/10.3390/ijms241612721
- Primary Citation of Related Structures:
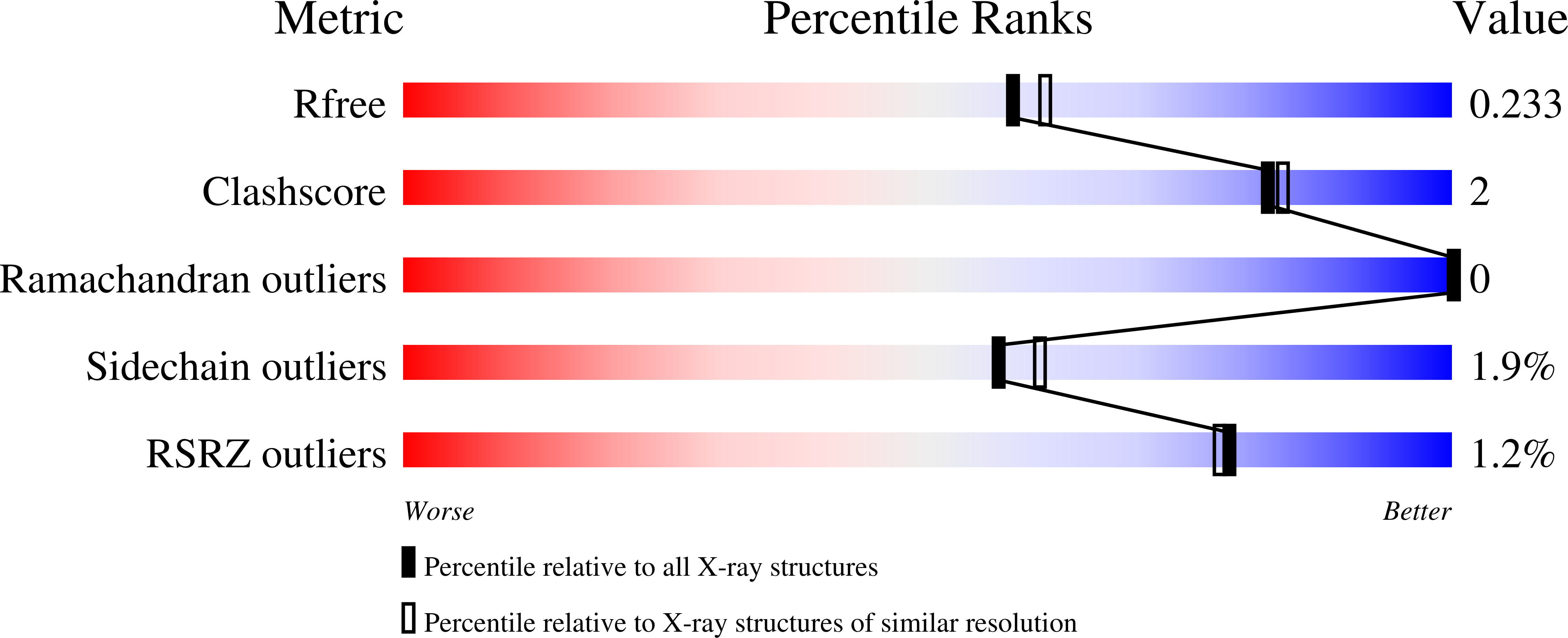
8OW8 - PubMed Abstract:
Clostridium botulinum neurotoxins (BoNTs) are the most potent toxins known, causing the deadly disease botulism. They function through Zn 2+ -dependent endopeptidase cleavage of SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins, preventing vesicular fusion and subsequent neurotransmitter release from motor neurons. Several serotypes of BoNTs produced by Clostridium botulinum (BoNT/A-/G and/X) have been well-characterised over the years. However, a BoNT-like gene (homologue of BoNT) was recently identified in the non-clostridial species, Enterococcus faecium, which is the leading cause of hospital-acquired multi-drug resistant infections. Here, we report the crystal structure of the catalytic domain of a BoNT homologue from Enterococcus faecium (LC/En) at 2.0 Å resolution. Detailed structural analysis in comparison with the full-length BoNT/En AlphaFold2-predicted structure, LC/A (from BoNT/A), and LC/F (from BoNT/F) revealed putative subsites and exosites (including loops 1-5) involved in recognition of LC/En substrates. LC/En also appears to possess a conserved autoproteolytic cleavage site whose function is yet to be established.
Organizational Affiliation:
Department of Life Sciences, University of Bath, Claverton Down, Bath BA2 7AY, UK.