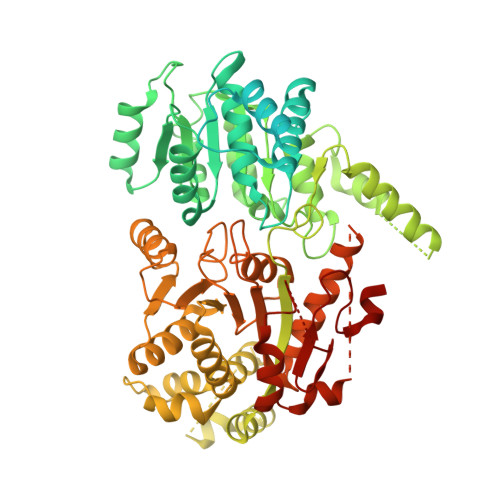
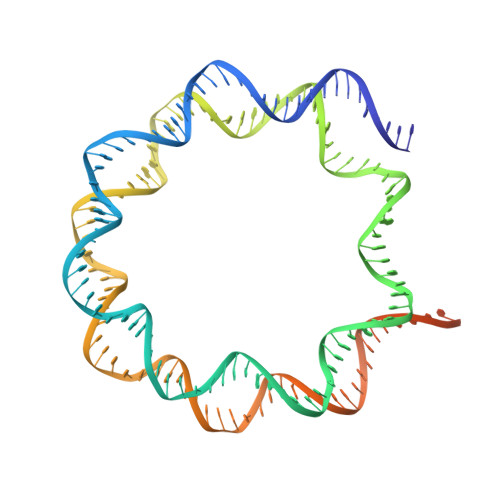
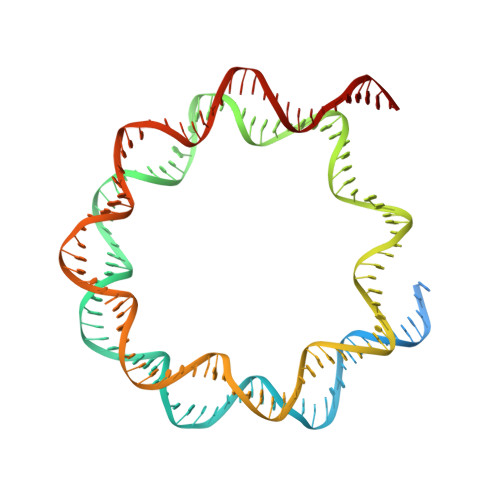
Mechanism of heterochromatin remodeling revealed by the DDM1 bound nucleosome structures.
Zhang, H., Gu, Z., Zeng, Y., Zhang, Y.(2024) Structure 32: 1222-1230.e4
- PubMed: 38870940
- DOI: https://doi.org/10.1016/j.str.2024.05.013
- Primary Citation of Related Structures:
8KCB, 8KCC - PubMed Abstract:
The SWI/SNF2 chromatin remodeling factor decreased DNA methylation 1 (DDM1) is essential for the silencing of transposable elements (TEs) in both euchromatic and heterochromatic regions. Here, we determined the cryo-EM structures of DDM1-nucleosome H2A and DDM1-nucleosome H2A.W complexes at near-atomic resolution in the presence of the ATP analog ADP-BeFx. The structures show that nucleosomal DNA is unwrapped more on the surface of the histone octamer containing histone H2A than that containing histone H2A.W. DDM1 embraces one DNA gyre of the nucleosome and interacts with the N-terminal tails of histone H4. Although we did not observe DDM1-H2A.W interactions in our structures, the results of the pull-down experiments suggest a direct interaction between DDM1 and the core region of histone H2A.W. Our work provides mechanistic insights into the heterochromatin remodeling process driven by DDM1 in plants.
Organizational Affiliation:
Key Laboratory of Synthetic Biology, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China. Electronic address: zhanghongwei@cemps.ac.cn.