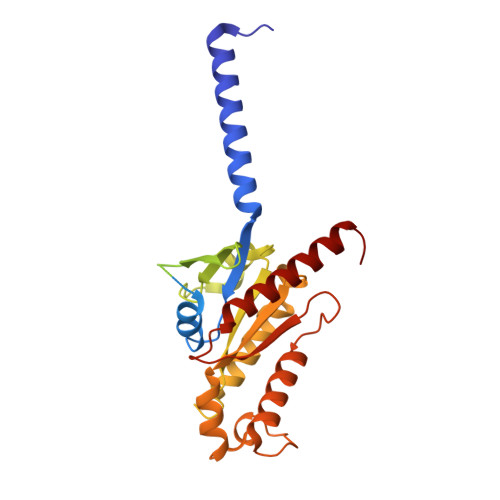
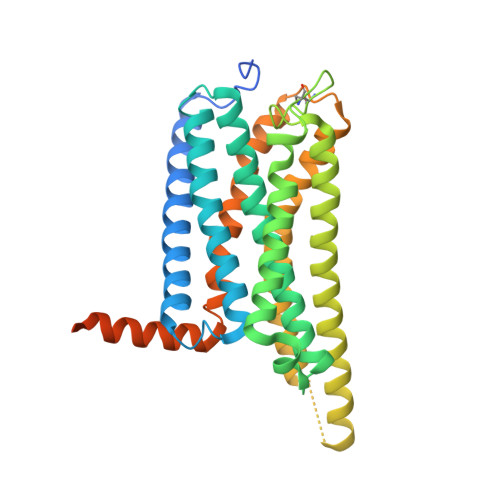
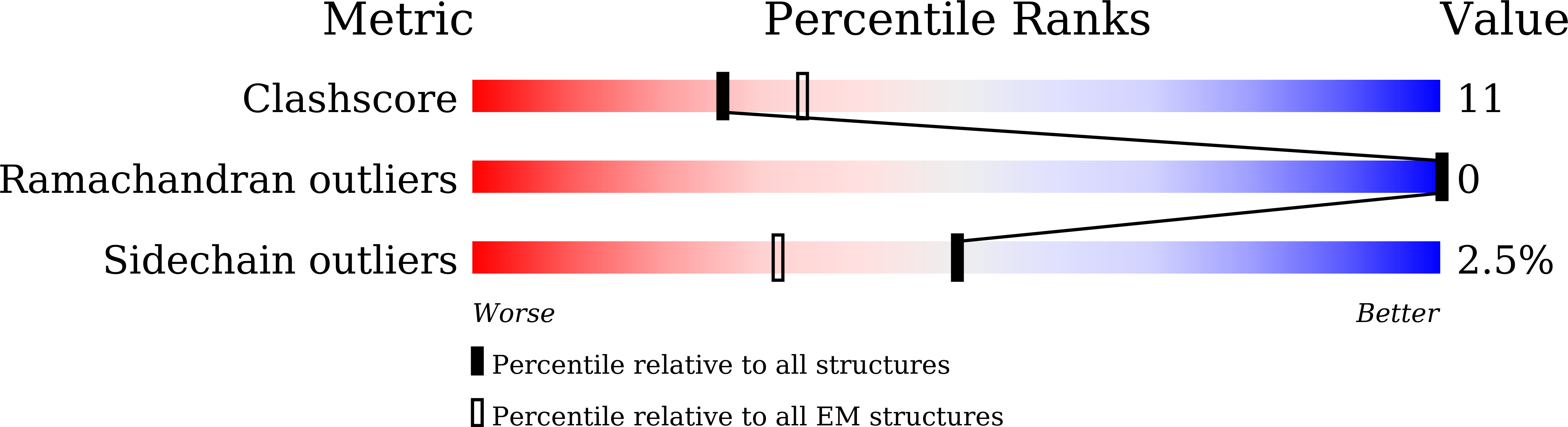Structural basis of selective cannabinoid CB 2 receptor activation.
Li, X., Chang, H., Bouma, J., de Paus, L.V., Mukhopadhyay, P., Paloczi, J., Mustafa, M., van der Horst, C., Kumar, S.S., Wu, L., Yu, Y., van den Berg, R.J.B.H.N., Janssen, A.P.A., Lichtman, A., Liu, Z.J., Pacher, P., van der Stelt, M., Heitman, L.H., Hua, T.(2023) Nat Commun 14: 1447-1447
- PubMed: 36922494
- DOI: https://doi.org/10.1038/s41467-023-37112-9
- Primary Citation of Related Structures:
8GUQ, 8GUR, 8GUS, 8GUT - PubMed Abstract:
Cannabinoid CB 2 receptor (CB 2 R) agonists are investigated as therapeutic agents in the clinic. However, their molecular mode-of-action is not fully understood. Here, we report the discovery of LEI-102, a CB 2 R agonist, used in conjunction with three other CBR ligands (APD371, HU308, and CP55,940) to investigate the selective CB 2 R activation by binding kinetics, site-directed mutagenesis, and cryo-EM studies. We identify key residues for CB 2 R activation. Highly lipophilic HU308 and the endocannabinoids, but not the more polar LEI-102, APD371, and CP55,940, reach the binding pocket through a membrane channel in TM1-TM7. Favorable physico-chemical properties of LEI-102 enable oral efficacy in a chemotherapy-induced nephropathy model. This study delineates the molecular mechanism of CB 2 R activation by selective agonists and highlights the role of lipophilicity in CB 2 R engagement. This may have implications for GPCR drug design and sheds light on their activation by endogenous ligands.
Organizational Affiliation:
iHuman Institute, ShanghaiTech University, Shanghai, 201210, China.