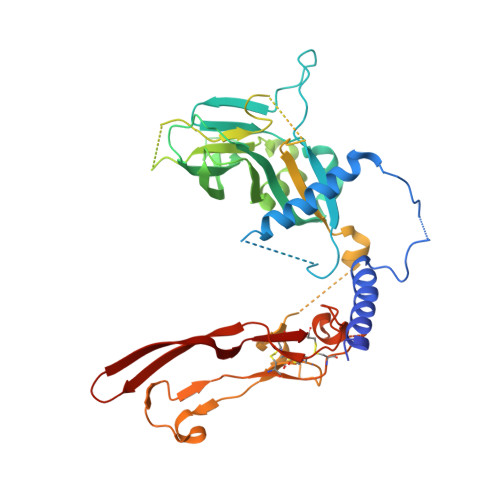
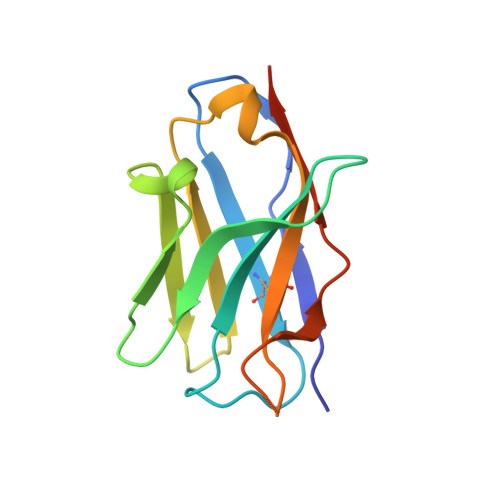
A specialized integrin-binding motif enables proTGF-beta 2 activation by integrin alpha V beta 6 but not alpha V beta 8.
Le, V.Q., Zhao, B., Ramesh, S., Toohey, C., DeCosta, A., Mintseris, J., Liu, X., Gygi, S., Springer, T.A.(2023) Proc Natl Acad Sci U S A 120: e2304874120-e2304874120
- PubMed: 37279271
- DOI: https://doi.org/10.1073/pnas.2304874120
- Primary Citation of Related Structures:
8FXS, 8FXV - PubMed Abstract:
Activation of latent transforming growth factor (TGF)-β2 is incompletely understood. Unlike TGF-β1 and β3, the TGF-β2 prodomain lacks a seven-residue RGDLXX (L/I) integrin-recognition motif and is thought not to be activated by integrins. Here, we report the surprising finding that TGF-β2 contains a related but divergent 13-residue integrin-recognition motif (YTSGDQKTIKSTR) that specializes it for activation by integrin αVβ6 but not αVβ8. Both classes of motifs compete for the same binding site in αVβ6. Multiple changes in the longer motif underlie its specificity. ProTGF-β2 structures define interesting differences from proTGF-β1 and the structural context for activation by αVβ6. Some integrin-independent activation is also seen for proTGF-β2 and even more so for proTGF-β3. Our findings have important implications for therapeutics to αVβ6 in clinical trials for fibrosis, in which inhibition of TGF-β2 activation has not been anticipated.
Organizational Affiliation:
Program in Cellular and Molecular Medicine, Department of Pediatrics, Boston Children's Hospital, Boston, MA 02115.