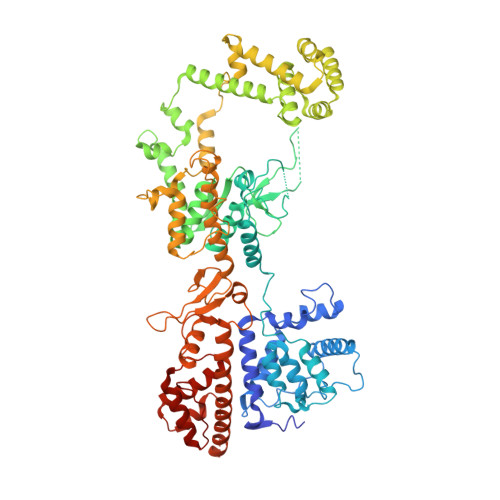
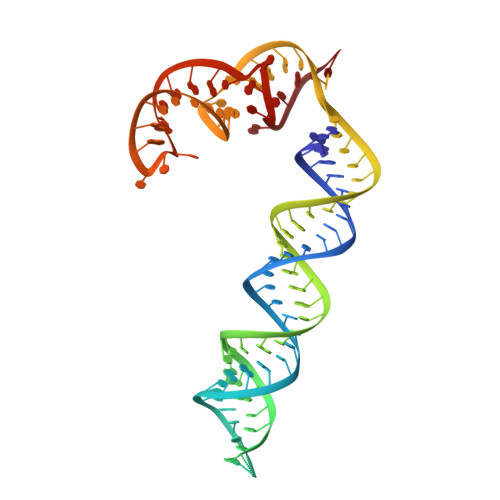
Structural basis for the activation of a compact CRISPR-Cas13 nuclease.
Deng, X., Osikpa, E., Yang, J., Oladeji, S.J., Smith, J., Gao, X., Gao, Y.(2023) Nat Commun 14: 5845-5845
- PubMed: 37730702
- DOI: https://doi.org/10.1038/s41467-023-41501-5
- Primary Citation of Related Structures:
8FTI - PubMed Abstract:
The CRISPR-Cas13 ribonucleases have been widely applied for RNA knockdown and transcriptional modulation owing to their high programmability and specificity. However, the large size of Cas13 effectors and their non-specific RNA cleavage upon target activation limit the adeno-associated virus based delivery of Cas13 systems for therapeutic applications. Herein, we report detailed biochemical and structural characterizations of a compact Cas13 (Cas13bt3) suitable for adeno-associated virus delivery. Distinct from many other Cas13 systems, Cas13bt3 cleaves the target and other nonspecific RNA at internal "UC" sites and is activated in a target length-dependent manner. The cryo-electron microscope structure of Cas13bt3 in a fully active state illustrates the structural basis of Cas13bt3 activation. Guided by the structure, we obtain engineered Cas13bt3 variants with minimal off-target cleavage yet maintained target cleavage activities. In conclusion, our biochemical and structural data illustrate a distinct mechanism for Cas13bt3 activation and guide the engineering of Cas13bt3 applications.
Organizational Affiliation:
Department of BioSciences, Rice University, Houston, TX, 77005, USA.