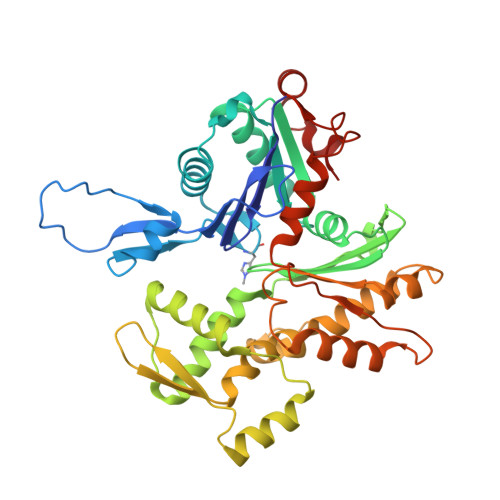
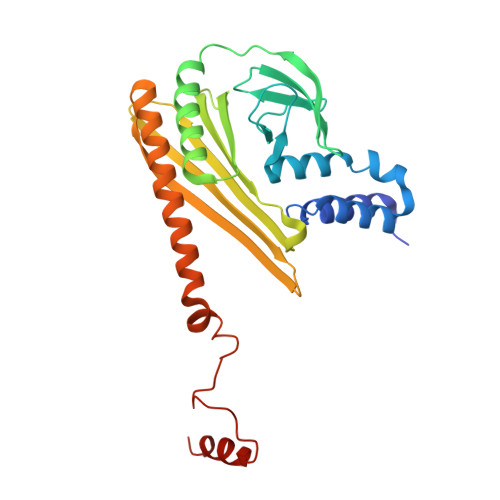
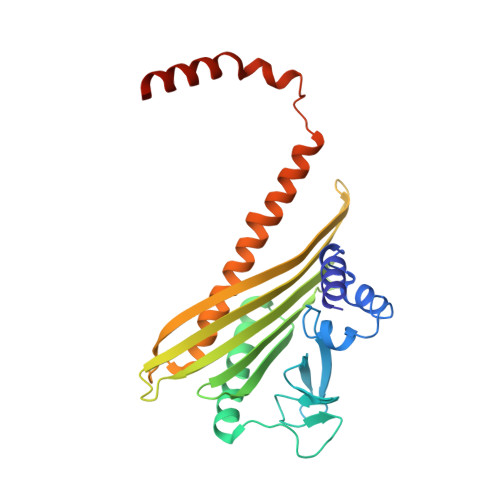
Structures of the free and capped ends of the actin filament.
Carman, P.J., Barrie, K.R., Rebowski, G., Dominguez, R.(2023) Science 380: 1287-1292
- PubMed: 37228182
- DOI: https://doi.org/10.1126/science.adg6812
- Primary Citation of Related Structures:
8F8P, 8F8Q, 8F8R, 8F8S, 8F8T - PubMed Abstract:
The barbed and pointed ends of the actin filament (F-actin) are the sites of growth and shrinkage and the targets of capping proteins that block subunit exchange, including CapZ at the barbed end and tropomodulin at the pointed end. We describe cryo-electron microscopy structures of the free and capped ends of F-actin. Terminal subunits at the free barbed end adopt a "flat" F-actin conformation. CapZ binds with minor changes to the barbed end but with major changes to itself. By contrast, subunits at the free pointed end adopt a "twisted" monomeric actin (G-actin) conformation. Tropomodulin binding forces the second subunit into an F-actin conformation. The structures reveal how the ends differ from the middle in F-actin and how these differences control subunit addition, dissociation, capping, and interactions with end-binding proteins.
Organizational Affiliation:
Department of Physiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.