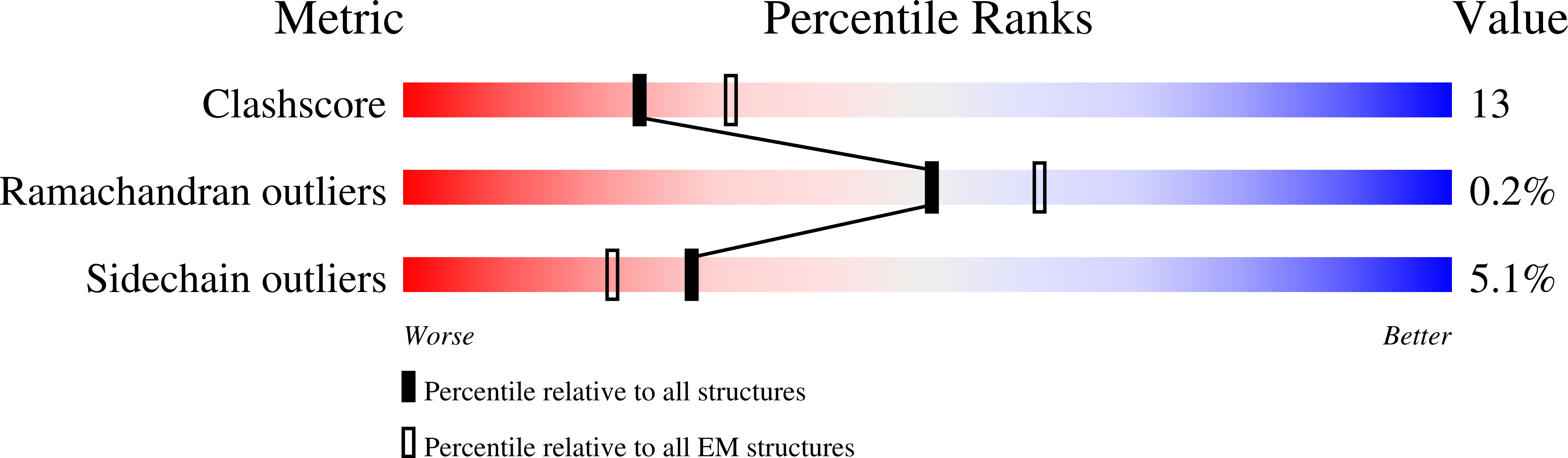Structural basis for membrane-proximal proteolysis of substrates by ADAM10.
Lipper, C.H., Egan, E.D., Gabriel, K.H., Blacklow, S.C.(2023) Cell 186: 3632-3641.e10
- PubMed: 37516108
- DOI: https://doi.org/10.1016/j.cell.2023.06.026
- Primary Citation of Related Structures:
8ESV - PubMed Abstract:
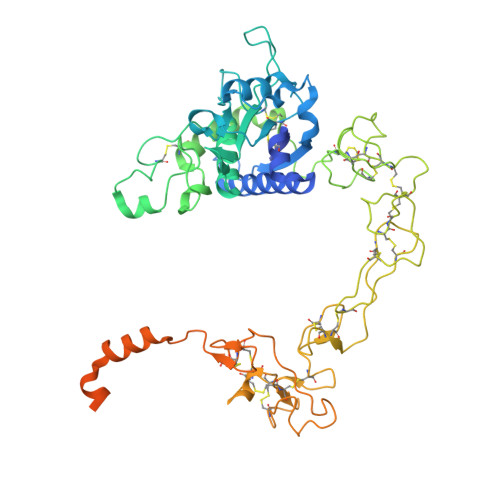
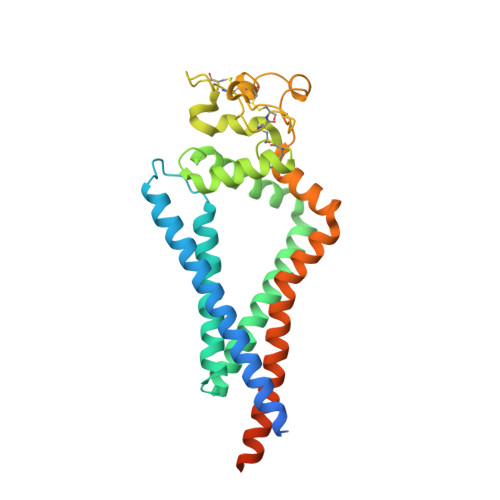
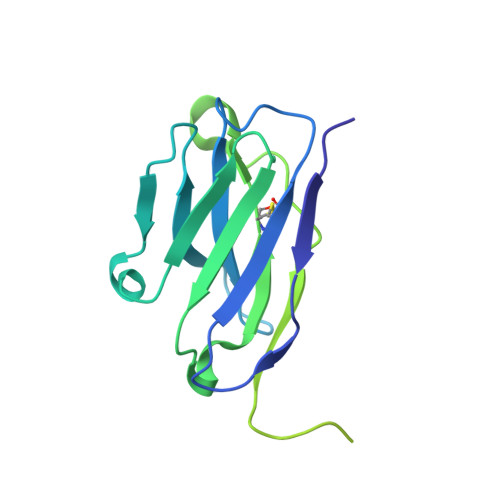
The endopeptidase ADAM10 is a critical catalyst for the regulated proteolysis of key drivers of mammalian development, physiology, and non-amyloidogenic cleavage of APP as the primary α-secretase. ADAM10 function requires the formation of a complex with a C8-tetraspanin protein, but how tetraspanin binding enables positioning of the enzyme active site for membrane-proximal cleavage remains unknown. We present here a cryo-EM structure of a vFab-ADAM10-Tspan15 complex, which shows that Tspan15 binding relieves ADAM10 autoinhibition and acts as a molecular measuring stick to position the enzyme active site about 20 Å from the plasma membrane for membrane-proximal substrate cleavage. Cell-based assays of N-cadherin shedding establish that the positioning of the active site by the interface between the ADAM10 catalytic domain and the bound tetraspanin influences selection of the preferred cleavage site. Together, these studies reveal the molecular mechanism underlying ADAM10 proteolysis at membrane-proximal sites and offer a roadmap for its modulation in disease.
Organizational Affiliation:
Department of Biological Chemistry and Molecular Pharmacology, Blavatnik Institute, Harvard Medical School, Boston, MA 02115, USA.