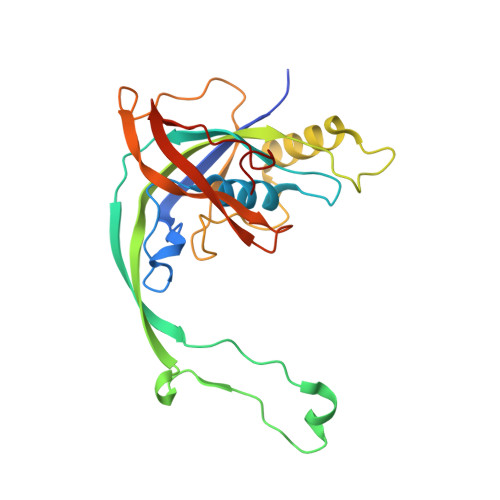
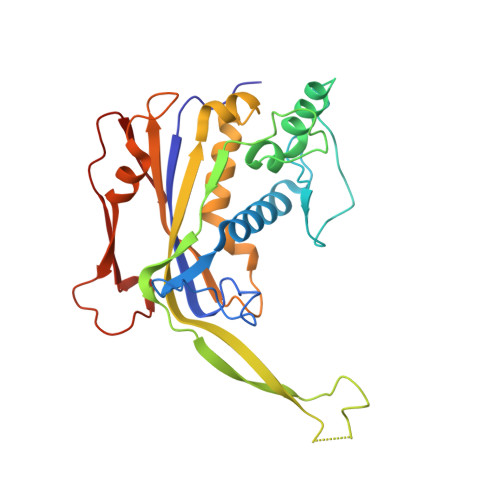
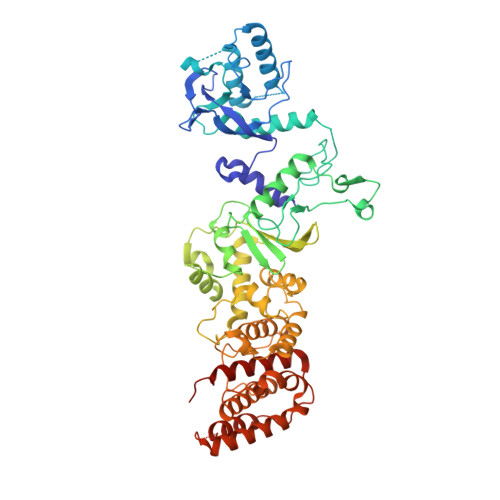
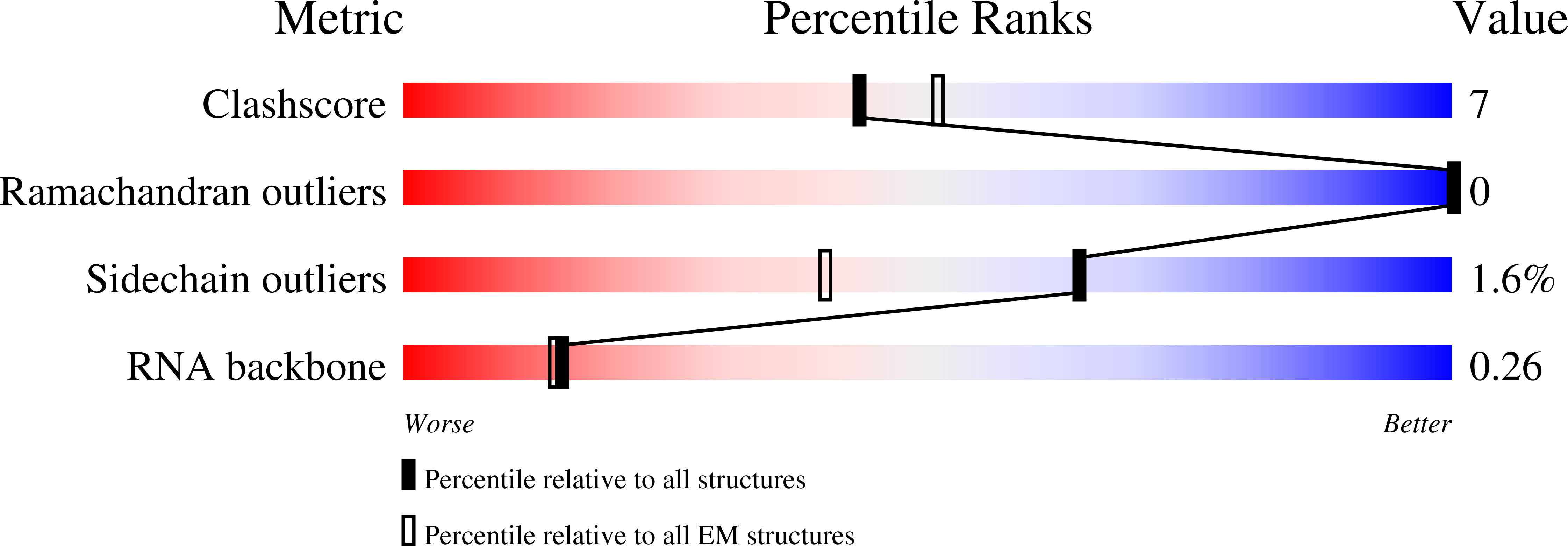Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
O'Brien, R.E., Bravo, J.P.K., Ramos, D., Hibshman, G.N., Wright, J.T., Taylor, D.W.(2023) Mol Cell 83: 746
- PubMed: 36805026
- DOI: https://doi.org/10.1016/j.molcel.2023.01.024
- Primary Citation of Related Structures:
8DEJ, 8DEX, 8DFA, 8DFO, 8DFS - PubMed Abstract:
Type I CRISPR-Cas systems employ multi-subunit Cascade effector complexes to target foreign nucleic acids for destruction. Here, we present structures of D. vulgaris type I-C Cascade at various stages of double-stranded (ds)DNA target capture, revealing mechanisms that underpin PAM recognition and Cascade allosteric activation. We uncover an interesting mechanism of non-target strand (NTS) DNA stabilization via stacking interactions with the "belly" subunits, securing the NTS in place. This "molecular seatbelt" mechanism facilitates efficient R-loop formation and prevents dsDNA reannealing. Additionally, we provide structural insights into how two anti-CRISPR (Acr) proteins utilize distinct strategies to achieve a shared mechanism of type I-C Cascade inhibition by blocking PAM scanning. These observations form a structural basis for directional R-loop formation and reveal how different Acr proteins have converged upon common molecular mechanisms to efficiently shut down CRISPR immunity.
Organizational Affiliation:
Interdisciplinary Life Sciences Graduate Programs, University of Texas at Austin, Austin, TX 78712, USA.