DNA-delivered antibody cocktail exhibits improved pharmacokinetics and confers prophylactic protection against SARS-CoV-2.
Parzych, E.M., Du, J., Ali, A.R., Schultheis, K., Frase, D., Smith, T.R.F., Cui, J., Chokkalingam, N., Tursi, N.J., Andrade, V.M., Warner, B.M., Gary, E.N., Li, Y., Choi, J., Eisenhauer, J., Maricic, I., Kulkarni, A., Chu, J.D., Villafana, G., Rosenthal, K., Ren, K., Francica, J.R., Wootton, S.K., Tebas, P., Kobasa, D., Broderick, K.E., Boyer, J.D., Esser, M.T., Pallesen, J., Kulp, D.W., Patel, A., Weiner, D.B.(2022) Nat Commun 13: 5886-5886
- PubMed: 36202799
- DOI: https://doi.org/10.1038/s41467-022-33309-6
- Primary Citation of Related Structures:
8D8Q, 8D8R - PubMed Abstract:
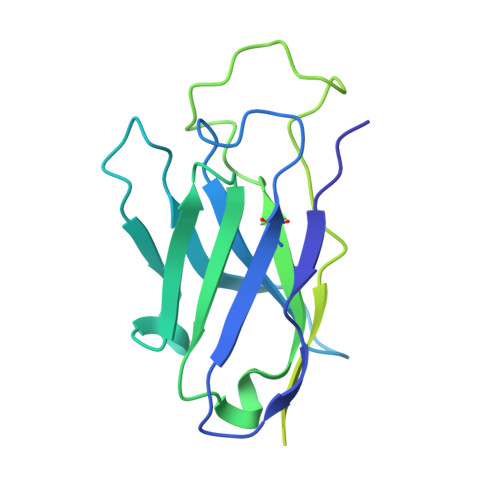
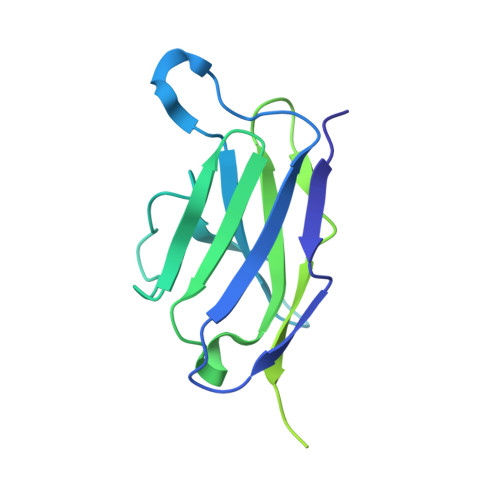
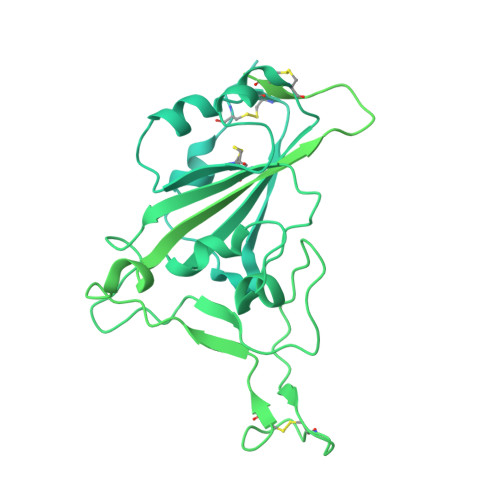
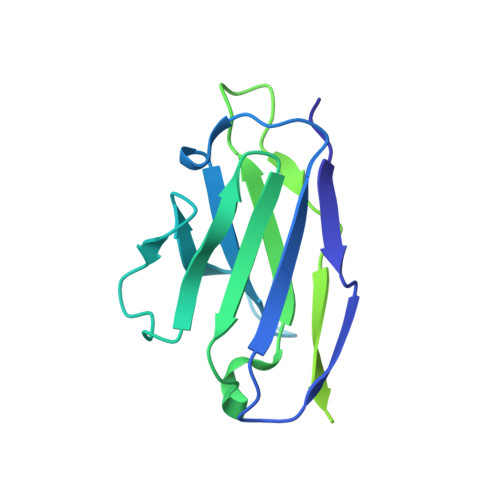
Monoclonal antibody therapy has played an important role against SARS-CoV-2. Strategies to deliver functional, antibody-based therapeutics with improved in vivo durability are needed to supplement current efforts and reach underserved populations. Here, we compare recombinant mAbs COV2-2196 and COV2-2130, which compromise clinical cocktail Tixagevimab/Cilgavimab, with optimized nucleic acid-launched forms. Functional profiling of in vivo-expressed, DNA-encoded monoclonal antibodies (DMAbs) demonstrated similar specificity, broad antiviral potency and equivalent protective efficacy in multiple animal challenge models of SARS-CoV-2 prophylaxis compared to protein delivery. In PK studies, DNA-delivery drove significant serum antibody titers that were better maintained compared to protein administration. Furthermore, cryo-EM studies performed on serum-derived DMAbs provide the first high-resolution visualization of in vivo-launched antibodies, revealing new interactions that may promote cooperative binding to trimeric antigen and broad activity against VoC including Omicron lineages. These data support the further study of DMAb technology in the development and delivery of valuable biologics.
Organizational Affiliation:
The Vaccine and Immunotherapy Center, The Wistar Institute of Anatomy and Biology, Philadelphia, PA, 19104, USA.