Structural atlas of the most abundant human gut virus
Bayfield, O.W., Shkoporov, A.N., Yutin, N., Khokhlova, E.V., Smith, J.L.R., Hawkins, D.E.D.P., Koonin, E.V., Hill, C., Antson, A.A.(null) Nature
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Currently 8CKB does not have a validation slider image.
(null) Nature
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Head fiber dimer protein gp29 | QS [auth A511] | 24 | Bacteroides phage crAss001 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A385DVL5 (Bacteroides phage crAss001) Explore A0A385DVL5 Go to UniProtKB: A0A385DVL5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A385DVL5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Muzzle bound helix | 14 | Bacteroides phage crAss001 | Mutation(s): 0 |  | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Muzzle protein gp44 | 1,371 | Bacteroides phage crAss001 | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for A0A385DVD6 (Bacteroides phage crAss001) Explore A0A385DVD6 Go to UniProtKB: A0A385DVD6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A385DVD6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cargo protein 1 gp45 | AWA [auth G008], BWA [auth G009], CWA [auth G010], DWA [auth G011], EWA [auth G012], FWA [auth G013], GWA [auth G014], HWA [auth G015], IWA [auth G016], JWA [auth G017], KWA [auth G018], LWA [auth G019], MWA [auth G020], NWA [auth G021], OWA [auth G022], PWA [auth G023], QWA [auth G024], TVA [auth G001], UVA [auth G002], VVA [auth G003], WVA [auth G004], XVA [auth G005], YVA [auth G006], ZVA [auth G007] | 842 | Bacteroides phage crAss001 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A385DV85 (Bacteroides phage crAss001) Explore A0A385DV85 Go to UniProtKB: A0A385DV85 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A385DV85 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 9 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Portal protein gp20 | 806 | Bacteroides phage crAss001 | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for A0A385DT68 (Bacteroides phage crAss001) Explore A0A385DT68 Go to UniProtKB: A0A385DT68 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A385DT68 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 10 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Portal vertex capsid protein gp57 | 104 | Bacteroides phage crAss001 | Mutation(s): 0 | 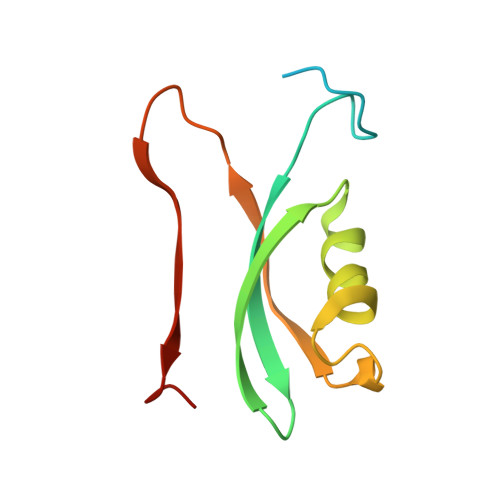 | |
UniProt | |||||
Find proteins for A0A385DTA3 (Bacteroides phage crAss001) Explore A0A385DTA3 Go to UniProtKB: A0A385DTA3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A385DTA3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 11 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ring protein 1 gp43 | 236 | Bacteroides phage crAss001 | Mutation(s): 0 | 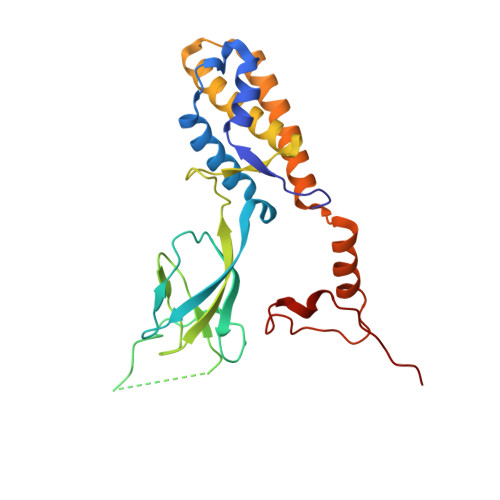 | |
UniProt | |||||
Find proteins for A0A385DT91 (Bacteroides phage crAss001) Explore A0A385DT91 Go to UniProtKB: A0A385DT91 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A385DT91 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 12 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ring protein 2 gp40 | 225 | Bacteroides phage crAss001 | Mutation(s): 0 | 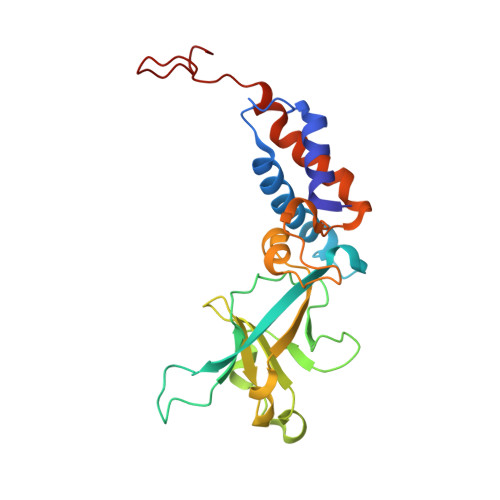 | |
UniProt | |||||
Find proteins for A0A385DT87 (Bacteroides phage crAss001) Explore A0A385DT87 Go to UniProtKB: A0A385DT87 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A385DT87 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 13 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ring protein 3 gp35 | 230 | Bacteroides phage crAss001 | Mutation(s): 0 | 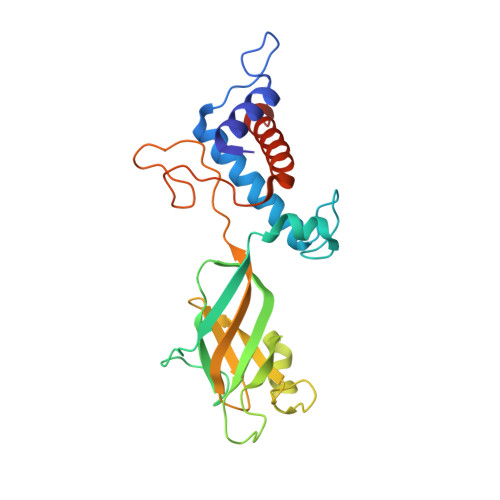 | |
UniProt | |||||
Find proteins for A0A385DV73 (Bacteroides phage crAss001) Explore A0A385DV73 Go to UniProtKB: A0A385DV73 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A385DV73 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 14 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ring protein 4/5 gp34 | AZA [auth M004], BZA [auth M005], CZA [auth M006], DZA [auth M007], EZA [auth M008], FZA [auth M009], GZA [auth M010], HZA [auth M011], IZA [auth M012], JZA [auth M013], KZA [auth M014], LZA [auth M015], MZA [auth M016], NZA [auth M017], OZA [auth M018], PZA [auth M019], QZA [auth M020], RZA [auth M021], SZA [auth M022], TZA [auth M023], UZA [auth M024], XYA [auth M001], YYA [auth M002], ZYA [auth M003] | 238 | Bacteroides phage crAss001 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A385DVC3 (Bacteroides phage crAss001) Explore A0A385DVC3 Go to UniProtKB: A0A385DVC3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A385DVC3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 15 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tail fiber protein gp22 | AAB [auth O006], ABB [auth O032], BAB [auth O007], BBB [auth O033], CAB [auth O008], CBB [auth O034], DAB [auth O009], DBB [auth O035], EAB [auth O010], EBB [auth O036], FAB [auth O011], GAB [auth O012], HAB [auth O013], IAB [auth O014], JAB [auth O015], KAB [auth O016], LAB [auth O017], MAB [auth O018], NAB [auth O019], OAB [auth O020], PAB [auth O021], QAB [auth O022], RAB [auth O023], SAB [auth O024], TAB [auth O025], UAB [auth O026], VAB [auth O027], VZA [auth O001], WAB [auth O028], WZA [auth O002], XAB [auth O029], XZA [auth O003], YAB [auth O030], YZA [auth O004], ZAB [auth O031], ZZA [auth O005] | 832 | Bacteroides phage crAss001 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A385DVT1 (Bacteroides phage crAss001) Explore A0A385DVT1 Go to UniProtKB: A0A385DVT1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A385DVT1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 16 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tail hub protein A gp38 | ACB [auth P022], BCB [auth P023], CCB [auth P024], FBB [auth P001], GBB [auth P002], HBB [auth P003], IBB [auth P004], JBB [auth P005], KBB [auth P006], LBB [auth P007], MBB [auth P008], NBB [auth P009], OBB [auth P010], PBB [auth P011], QBB [auth P012], RBB [auth P013], SBB [auth P014], TBB [auth P015], UBB [auth P016], VBB [auth P017], WBB [auth P018], XBB [auth P019], YBB [auth P020], ZBB [auth P021] | 215 | Bacteroides phage crAss001 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A385DTH1 (Bacteroides phage crAss001) Explore A0A385DTH1 Go to UniProtKB: A0A385DTH1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A385DTH1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 17 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tail hub protein B gp39 | 114 | Bacteroides phage crAss001 | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for A0A385DVM6 (Bacteroides phage crAss001) Explore A0A385DVM6 Go to UniProtKB: A0A385DVM6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A385DVM6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MG Query on MG | ADB [auth E120] AEB [auth E354] AFB [auth E508] BDB [auth E129] BEB [auth E363] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
Currently 8CKB does not have a validation slider image.
| Funding Organization | Location | Grant Number |
|---|---|---|
| Wellcome Trust | United Kingdom | 206377 |