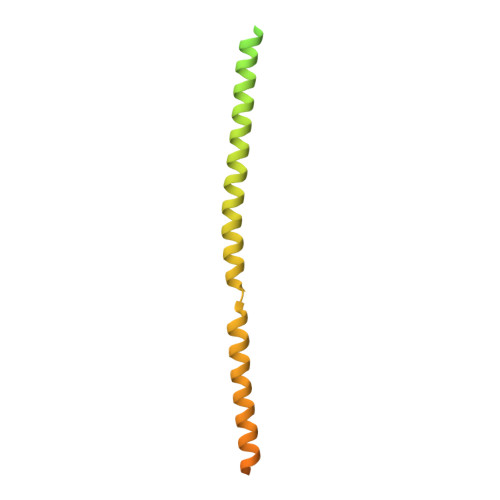Structural and biophysical characterization of the Borna disease virus 1 phosphoprotein.
Whitehead, J.D., Grimes, J.M., Keown, J.R.(2023) Acta Crystallogr F Struct Biol Commun 79: 51-60
- PubMed: 36862093
- DOI: https://doi.org/10.1107/S2053230X23000717
- Primary Citation of Related Structures:
8BS7 - PubMed Abstract:
Bornaviruses are RNA viruses with a mammalian, reptilian, and avian host range. The viruses infect neuronal cells and in rare cases cause a lethal encephalitis. The family Bornaviridae are part of the Mononegavirales order of viruses, which contain a nonsegmented viral genome. Mononegavirales encode a viral phosphoprotein (P) that binds both the viral polymerase (L) and the viral nucleoprotein (N). The P protein acts as a molecular chaperone and is required for the formation of a functional replication/transcription complex. In this study, the structure of the oligomerization domain of the phosphoprotein determined by X-ray crystallography is reported. The structural results are complemented with biophysical characterization using circular dichroism, differential scanning calorimetry and small-angle X-ray scattering. The data reveal the phosphoprotein to assemble into a stable tetramer, with the regions outside the oligomerization domain remaining highly flexible. A helix-breaking motif is observed between the α-helices at the midpoint of the oligomerization domain that appears to be conserved across the Bornaviridae. These data provide information on an important component of the bornavirus replication complex.
Organizational Affiliation:
Division of Structural Biology, Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, United Kingdom.