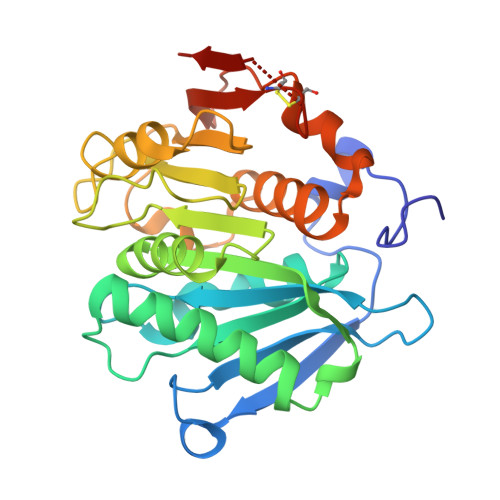
The metagenome-derived esterase PET40 is highly promiscuous and hydrolyses polyethylene terephthalate (PET).
Zhang, H., Dierkes, R.F., Perez-Garcia, P., Costanzi, E., Dittrich, J., Cea, P.A., Gurschke, M., Applegate, V., Partus, K., Schmeisser, C., Pfleger, C., Gohlke, H., Smits, S.H.J., Chow, J., Streit, W.R.(2024) FEBS J 291: 70-91
- PubMed: 37549040
- DOI: https://doi.org/10.1111/febs.16924
- Primary Citation of Related Structures:
8A2C - PubMed Abstract:
Polyethylene terephthalate (PET) is a widely used synthetic polymer and known to contaminate marine and terrestrial ecosystems. Only few PET-active microorganisms and enzymes (PETases) are currently known, and it is debated whether degradation activity for PET originates from promiscuous enzymes with broad substrate spectra that primarily act on natural polymers or other bulky substrates, or whether microorganisms evolved their genetic makeup to accepting PET as a carbon source. Here, we present a predicted diene lactone hydrolase designated PET40, which acts on a broad spectrum of substrates, including PET. It is the first esterase with activity on PET from a GC-rich Gram-positive Amycolatopsis species belonging to the Pseudonocardiaceae (Actinobacteria). It is highly conserved within the genera Amycolatopsis and Streptomyces. PET40 was identified by sequence-based metagenome search using a PETase-specific hidden Markov model. Besides acting on PET, PET40 has a versatile substrate spectrum, hydrolyzing δ-lactones, β-lactam antibiotics, the polyester-polyurethane Impranil® DLN, and various para-nitrophenyl ester substrates. Molecular docking suggests that the PET degradative activity is likely a result of the promiscuity of PET40, as potential binding modes were found for substrates encompassing mono(2-hydroxyethyl) terephthalate, bis(2-hydroxyethyl) terephthalate, and a PET trimer. We also solved the crystal structure of the inactive PET40 variant S178A to 1.60 Å resolution. PET40 is active throughout a wide pH (pH 4-10) and temperature range (4-65 °C) and remarkably stable in the presence of 5% SDS, making it a promising enzyme as a starting point for further investigations and optimization approaches.
Organizational Affiliation:
Department of Microbiology and Biotechnology, University of Hamburg, Germany.