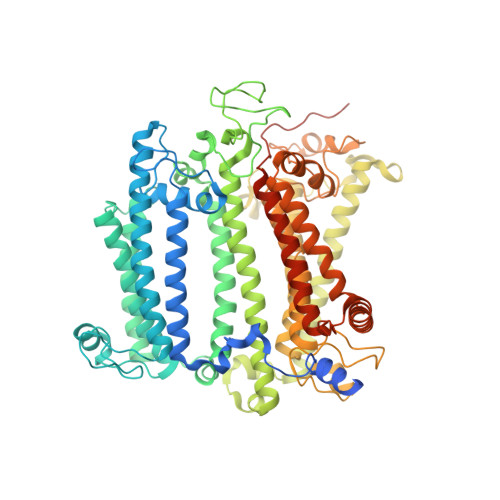
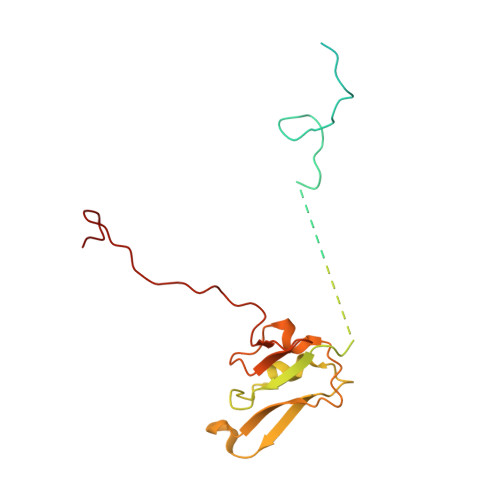
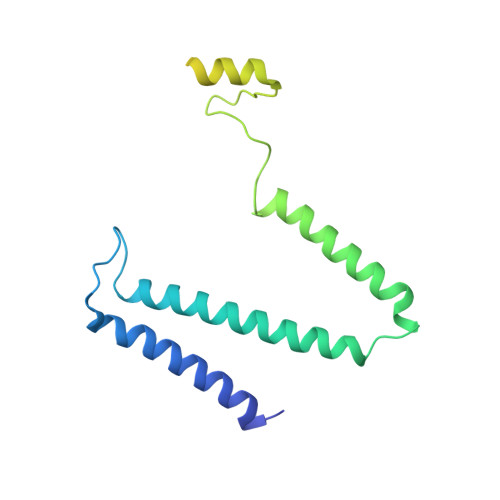
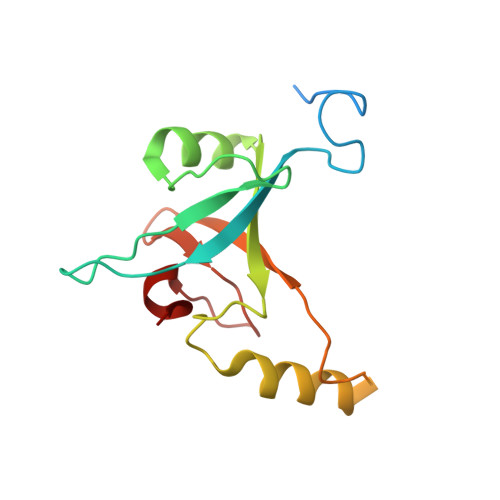
Cryo-EM structure of the whole photosynthetic reaction center apparatus from the green sulfur bacterium Chlorobaculum tepidum.
Xie, H., Lyratzakis, A., Khera, R., Koutantou, M., Welsch, S., Michel, H., Tsiotis, G.(2023) Proc Natl Acad Sci U S A 120: e2216734120-e2216734120
- PubMed: 36693097
- DOI: https://doi.org/10.1073/pnas.2216734120
- Primary Citation of Related Structures:
7Z6Q - PubMed Abstract:
Light energy absorption and transfer are very important processes in photosynthesis. In green sulfur bacteria light is absorbed primarily by the chlorosomes and its energy is transferred via the Fenna-Matthews-Olson (FMO) proteins to a homodimeric reaction center (RC). Here, we report the cryogenic electron microscopic structure of the intact FMO-RC apparatus from Chlorobaculum tepidum at 2.5 Å resolution. The FMO-RC apparatus presents an asymmetric architecture and contains two FMO trimers that show different interaction patterns with the RC core. Furthermore, the two permanently bound transmembrane subunits PscC, which donate electrons to the special pair, interact only with the two large PscA subunits. This structure fills an important gap in our understanding of the transfer of energy from antenna to the electron transport chain of this RC and the transfer of electrons from reduced sulfur compounds to the special pair.
Organizational Affiliation:
Department of Molecular Membrane Biology, Max Planck Institute of Biophysics, Frankfurt am Main D-60438, Germany.