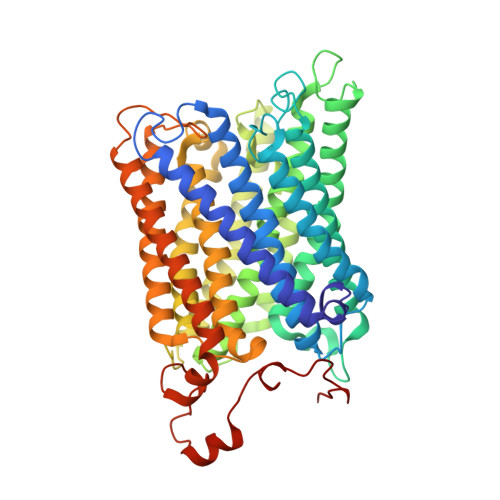
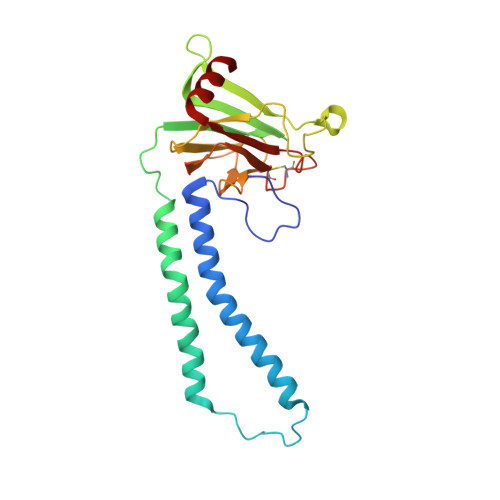
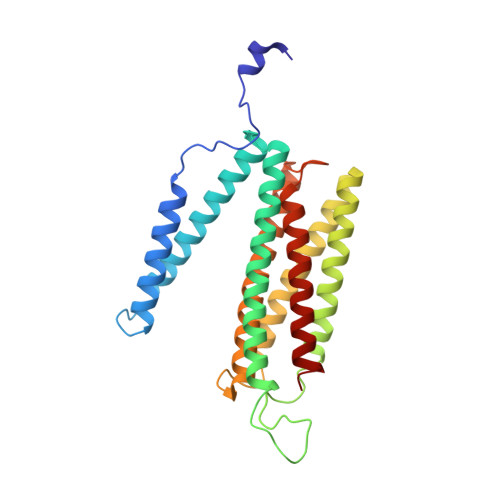
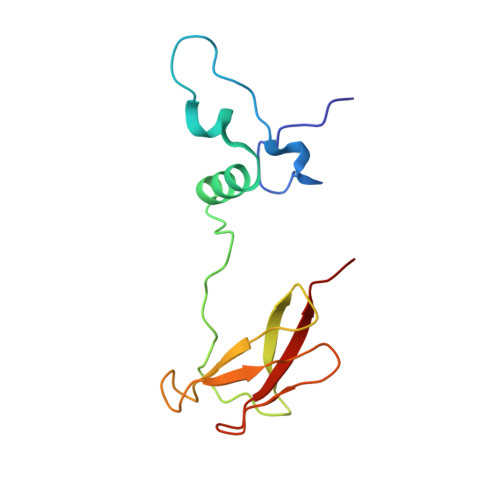
Cryo-EM structure of a monomeric yeast S. cerevisiae complex IV isolated with maltosides: Implications in supercomplex formation.
Ing, G., Hartley, A.M., Pinotsis, N., Marechal, A.(2022) Biochim Biophys Acta Bioenerg 1863: 148591-148591
- PubMed: 35839926
- DOI: https://doi.org/10.1016/j.bbabio.2022.148591
- Primary Citation of Related Structures:
7Z10 - PubMed Abstract:
In mitochondria, complex IV (CIV) can be found as a monomer, a dimer or in association with other respiratory complexes. The atomic structure of the yeast S. cerevisiae CIV in a supercomplex (SC) with complex III (CIII) pointed to a region of significant conformational changes compared to the homologous mammalian CIV structures. These changes involved the matrix side domain of Cox5A at the CIII-CIV interface, and it was suggested that it could be required for SC formation. To investigate this, we solved the structure of the isolated monomeric CIV from S. cerevisiae stabilised in amphipol A8-35 at 3.9 Å using cryo-electron microscopy. Only a minor change in flexibility was seen in this Cox5A region, ruling out large CIV conformational shift for interaction with CIII and confirming the different fold of the yeast Cox5A subunit compared to mammalian homologues. Other differences in structure were the absence of two canonical subunits, Cox12 and Cox13, as well as Cox26, which is unique to the yeast CIV. Their absence is most likely due to the protein purification protocol used to isolate CIV from the III-IV SC.
Organizational Affiliation:
Institute of Structural and Molecular Biology, University College London, Gower Street, London WC1E 6BT, UK.