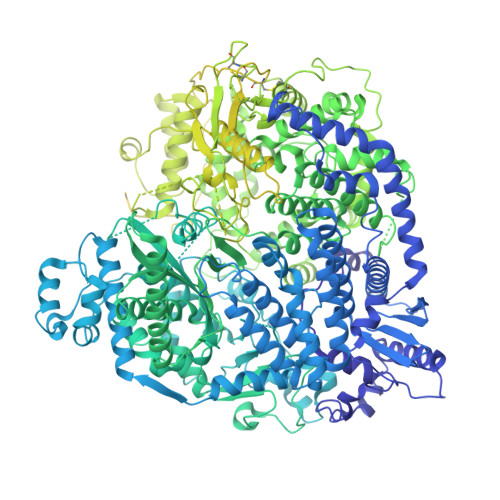
Structure of the Newcastle Disease Virus L protein in complex with tetrameric phosphoprotein.
Cong, J., Feng, X., Kang, H., Fu, W., Wang, L., Wang, C., Li, X., Chen, Y., Rao, Z.(2023) Nat Commun 14: 1324-1324
- PubMed: 36898997
- DOI: https://doi.org/10.1038/s41467-023-37012-y
- Primary Citation of Related Structures:
7YOT, 7YOU, 7YOV - PubMed Abstract:
Newcastle disease virus (NDV) belongs to Paramyxoviridae, which contains lethal human and animal pathogens. NDV RNA genome is replicated and transcribed by a multifunctional 250 kDa RNA-dependent RNA polymerase (L protein). To date, high-resolution structure of NDV L protein complexed with P protein remains to be elucidated, limiting our understanding of the molecular mechanisms of Paramyxoviridae replication/transcription. Here, we used cryo-EM and enzymatic assays to investigate the structure-function relationship of L-P complex. We found that C-terminal of CD-MTase-CTD module of the atomic-resolution L-P complex conformationally rearranges, and the priming/intrusion loops are likely in RNA elongation conformations different from previous structures. The P protein adopts a unique tetrameric organization and interacts with L protein. Our findings indicate that NDV L-P complex represents elongation state distinct from previous structures. Our work greatly advances the understanding of Paramyxoviridae RNA synthesis, revealing how initiation/elongation alternates, providing clues for identifying therapeutic targets against Paramyxoviridae.
Organizational Affiliation:
National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China.