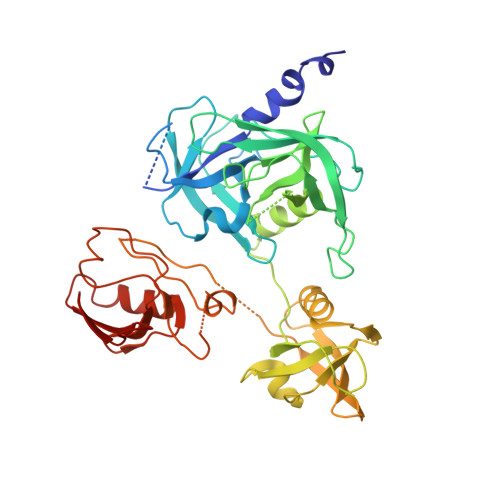
Crystal structures and solution conformations of HtrA from Helicobacter pylori reveal pH-dependent oligomeric conversion and conformational rearrangements.
Cui, L., Shi, X., Li, H., Wang, S., Guo, L., Lan, Z., Dai, Y., Zhang, Q., Wu, Y., Liu, W.(2023) Int J Biol Macromol 243: 125274-125274
- PubMed: 37301353
- DOI: https://doi.org/10.1016/j.ijbiomac.2023.125274
- Primary Citation of Related Structures:
7XS0, 7XS2 - PubMed Abstract:
Helicobacter pylori is a Gram-negative microaerophilic bacterium that infects over 50 % of the world's population, making it a major risk factor for chronic gastritis, ulcer diseases of the stomach and duodenum, MALT lymphoma, and gastric cancer. The clinical consequences of H. pylori infection are closely linked with the expression of virulence factors secreted by the bacterium. One such virulence factor is high temperature requirement A (HtrA), which possesses chaperone and serine protease activity. In the host stomach, HtrA secreted from H. pylori (HpHtrA) disrupts intercellular adhesions by cleaving epithelial adhesion proteins including E-cadherin and desmoglein-2. This disruption causes intercellular junctions to open, allowing the bacterium to pass through the epithelial barrier, access the intercellular space, and colonize the gastric mucosa. HtrA proteases are well known for their structural complexity, reflected in their diverse oligomer forms and multi-tasking activities in both prokaryotes and eukaryotes. In this study, we determined crystal structures and solution conformations of HpHtrA monomer and trimer, which revealed large domain rearrangements between them. Notably, this is the first report of a monomeric structure in the HtrA family. We further found a pH-dependent dynamic trimer-to-monomer conversion and concurrent conformational changes that seem closely linked with a pH-sensing ability through the protonation of certain Asp residues. These results advance our understanding of the functional roles and the related mechanisms of this protease in bacterial infection, which may shed light on the development of HtrA-targeted therapies for H. pylori-associated diseases.
Organizational Affiliation:
Institute of Immunology, PLA, Army Medical University, Chongqing 400038, China; Department of Tropical Medicine and Infectious Diseases, Hainan Hospital of Chinese PLA General Hospital, Sanya, Hainan 572000, China.