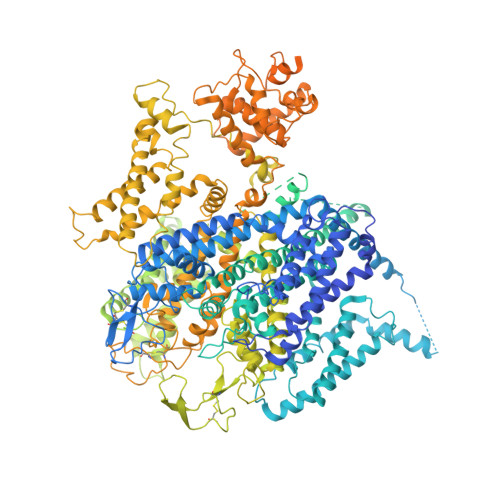
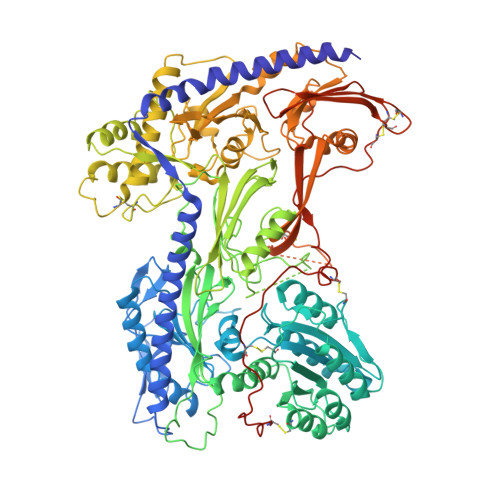
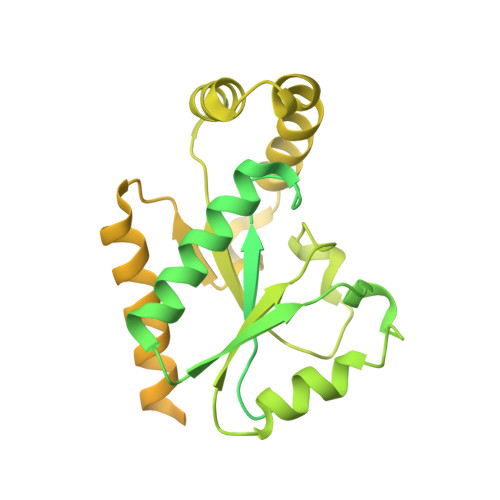
Molecular insights into the gating mechanisms of voltage-gated calcium channel Ca V 2.3.
Gao, Y., Xu, S., Cui, X., Xu, H., Qiu, Y., Wei, Y., Dong, Y., Zhu, B., Peng, C., Liu, S., Zhang, X.C., Sun, J., Huang, Z., Zhao, Y.(2023) Nat Commun 14: 516-516
- PubMed: 36720859
- DOI: https://doi.org/10.1038/s41467-023-36260-2
- Primary Citation of Related Structures:
7XLQ - PubMed Abstract:
High-voltage-activated R-type Ca V 2.3 channel plays pivotal roles in many physiological activities and is implicated in epilepsy, convulsions, and other neurodevelopmental impairments. Here, we determine the high-resolution cryo-electron microscopy (cryo-EM) structure of human Ca V 2.3 in complex with the α2δ1 and β1 subunits. The VSD II is stabilized in the resting state. Electrophysiological experiments elucidate that the VSD II is not required for channel activation, whereas the other VSDs are essential for channel opening. The intracellular gate is blocked by the W-helix. A pre-W-helix adjacent to the W-helix can significantly regulate closed-state inactivation (CSI) by modulating the association and dissociation of the W-helix with the gate. Electrostatic interactions formed between the negatively charged domain on S6 II , which is exclusively conserved in the Ca V 2 family, and nearby regions at the alpha-interacting domain (AID) and S4-S5 II helix are identified. Further functional analyses indicate that these interactions are critical for the open-state inactivation (OSI) of Ca V 2 channels.
Organizational Affiliation:
National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China.