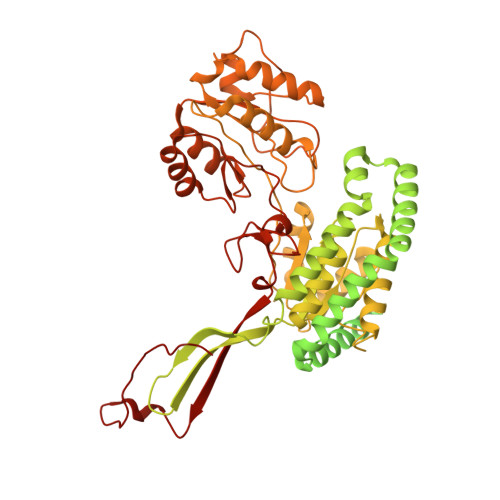
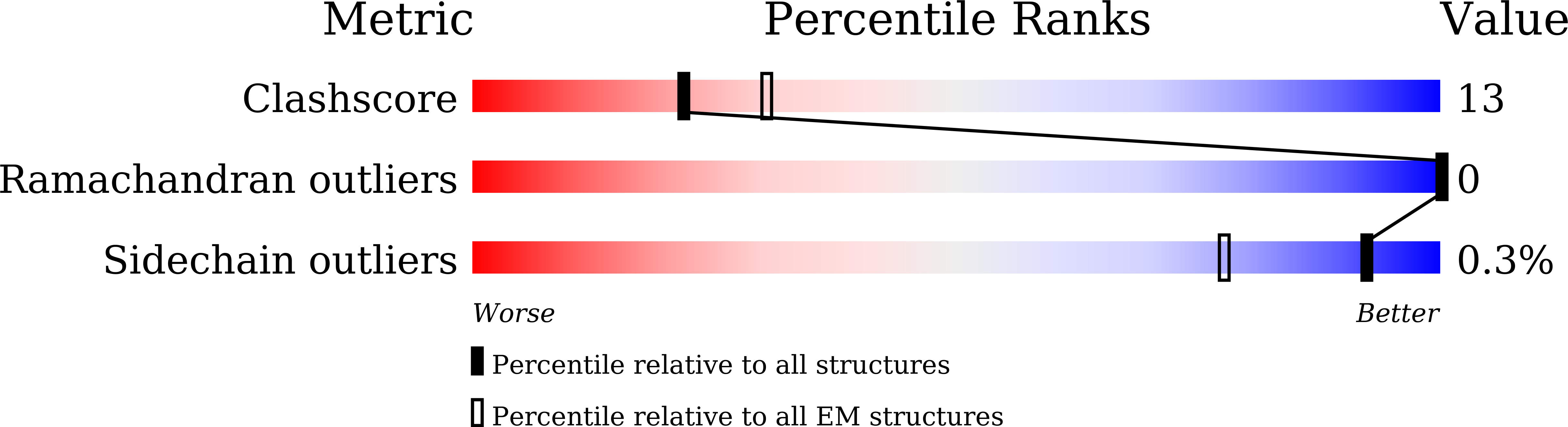Structural basis of dynamic P5CS filaments.
Zhong, J., Guo, C.J., Zhou, X., Chang, C.C., Yin, B., Zhang, T., Hu, H., Lu, G.M., Liu, J.L.(2022) Elife 11
- PubMed: 35286254
- DOI: https://doi.org/10.7554/eLife.76107
- Primary Citation of Related Structures:
7F5T, 7F5U, 7F5V, 7F5X, 7WX3, 7WX4, 7WXF, 7WXG, 7WXH, 7WXI - PubMed Abstract:
The bifunctional enzyme Δ 1 -pyrroline-5-carboxylate synthase (P5CS) is vital to the synthesis of proline and ornithine, playing an essential role in human health and agriculture. Pathogenic mutations in the P5CS gene (ALDH18A1) lead to neurocutaneous syndrome and skin relaxation connective tissue disease in humans, and P5CS deficiency seriously damages the ability to resist adversity in plants. We have recently found that P5CS forms cytoophidia in vivo and filaments in vitro. However, it is difficult to appreciate the function of P5CS filamentation without precise structures. Using cryo-electron microscopy, here we solve the structures of Drosophila full-length P5CS in three states at resolution from 3.1 to 4.3 Å. We observe distinct ligand-binding states and conformational changes for the GK and GPR domains, respectively. Divergent helical filaments are assembled by P5CS tetramers and stabilized by multiple interfaces. Point mutations disturbing those interfaces prevent P5CS filamentation and greatly reduce the enzymatic activity. Our findings reveal that filamentation is crucial for the coordination between the GK and GPR domains, providing a structural basis for the catalytic function of P5CS filaments.
Organizational Affiliation:
School of Life Science and Technology, ShanghaiTech University, Shanghai, China.