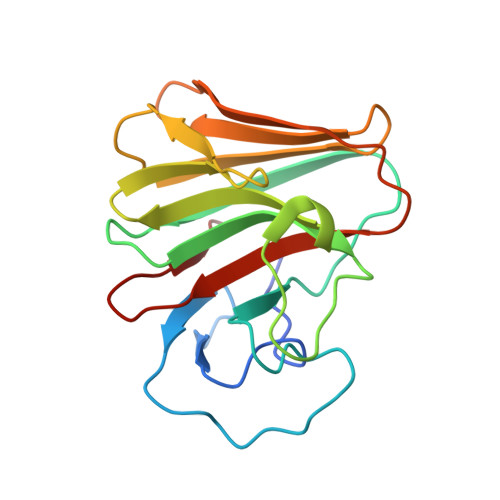
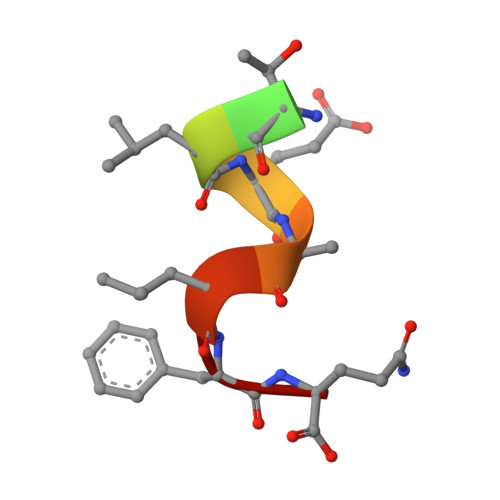
A C-terminal glutamine recognition mechanism revealed by E3 ligase TRIM7 structures.
Liang, X., Xiao, J., Li, X., Liu, Y., Lu, Y., Wen, Y., Li, Z., Che, X., Ma, Y., Zhang, X., Zhang, Y., Jian, D., Wang, P., Xuan, C., Yu, G., Li, L., Zhang, H.(2022) Nat Chem Biol 18: 1214-1223
- PubMed: 35982226
- DOI: https://doi.org/10.1038/s41589-022-01128-x
- Primary Citation of Related Structures:
7W0Q, 7W0S, 7W0T, 7X6Y, 7X6Z, 7X70 - PubMed Abstract:
The E3 ligase TRIM7 has emerged as a critical player in viral infection and pathogenesis. However, the mechanism governing the TRIM7-substrate association remains to be defined. Here we report the crystal structures of TRIM7 in complex with 2C peptides of human enterovirus. Structure-guided studies reveal the C-terminal glutamine residue of 2C as the primary determinant for TRIM7 binding. Leveraged by this finding, we identify norovirus and SARS-CoV-2 proteins, and physiological proteins, as new TRIM7 substrates. Crystal structures of TRIM7 in complex with multiple peptides derived from SARS-CoV-2 proteins display the same glutamine-end recognition mode. Furthermore, TRIM7 could trigger the ubiquitination and degradation of these substrates, possibly representing a new Gln/C-degron pathway. Together, these findings unveil a common recognition mode by TRIM7, providing the foundation for further mechanistic characterization of antiviral and cellular functions of TRIM7.
Organizational Affiliation:
Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Haihe Laboratory of Cell Ecosystem, Tianjin Institute of Immunology, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin, China.