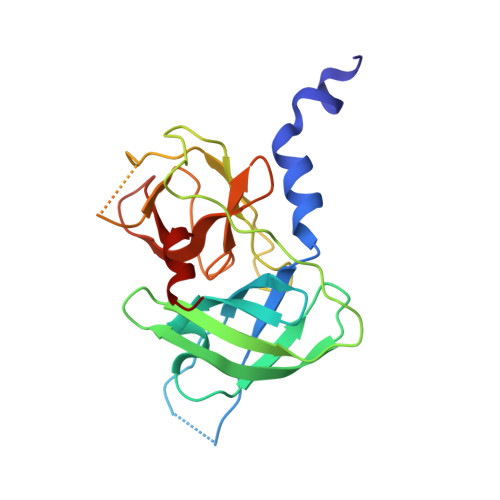
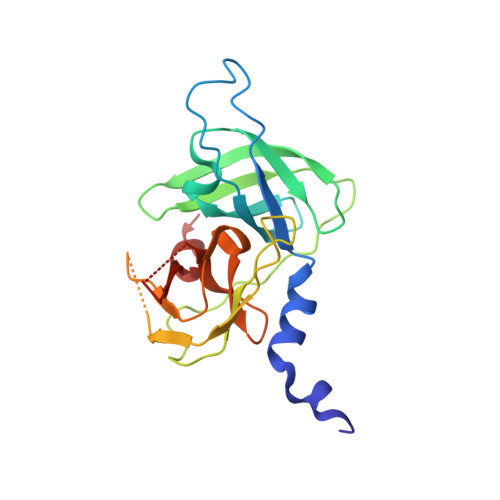
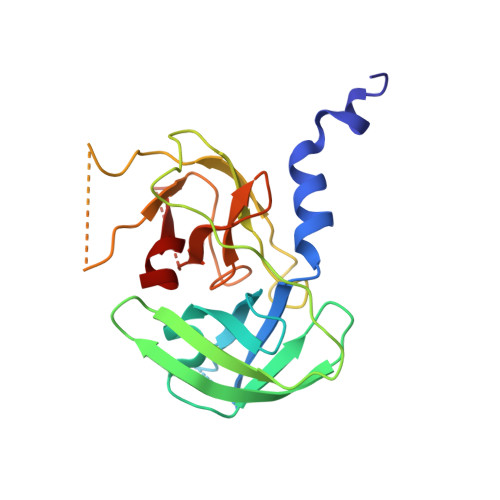
Inter-subunit crosstalk via PDZ synergistically governs allosteric activation of proapoptotic HtrA2.
Parui, A.L., Mishra, V., Dutta, S., Bhaumik, P., Bose, K.(2022) Structure 30: 1307-1320.e5
- PubMed: 35738282
- DOI: https://doi.org/10.1016/j.str.2022.06.001
- Primary Citation of Related Structures:
7VGE - PubMed Abstract:
The mitochondrial serine protease High-temperature requirement A2 (HtrA2) is associated with various diseases including neurodegenerative disorders and cancer. Despite availability of structural details, the reports on HtrA2's mechanistic regulation that varies with the type of activation signals still remain non-concordant. To expound the role of regulatory PDZ (Postsynaptic density-95/Discs large/Zonula occludens-1) domains in multimodal activation of HtrA2, we generated heterotrimeric HtrA2 variants comprising different numbers of PDZs and/or active-site mutations. Sequential deletion of PDZs from the trimeric ensemble significantly affected its residual activity in a way that proffered a hypothesis advocating inter-molecular allosteric crosstalk via PDZs in HtrA2. Furthermore, structural and computational snapshots affirmed the role of PDZs in secondary structural element formation around the regulatory loops and coordinated reorganization of the N-terminal region. Therefore, apart from providing cues for devising structure-guided therapeutic strategies, this study establishes a physiologically relevant working model of complex allosteric regulation through a trans-mediated cooperatively shared energy landscape.
Organizational Affiliation:
Integrated Biophysics and Structural Biology Lab, ACTREC - Tata Memorial Centre, Navi Mumbai 410210, India; Homi Bhabha National Institute, BARC Training School Complex, Anushaktinagar, Mumbai 400094, India.