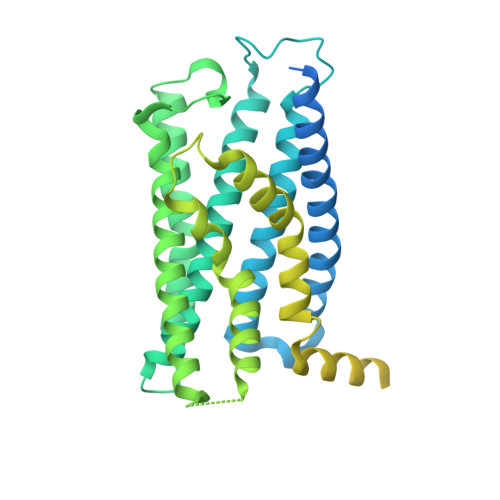
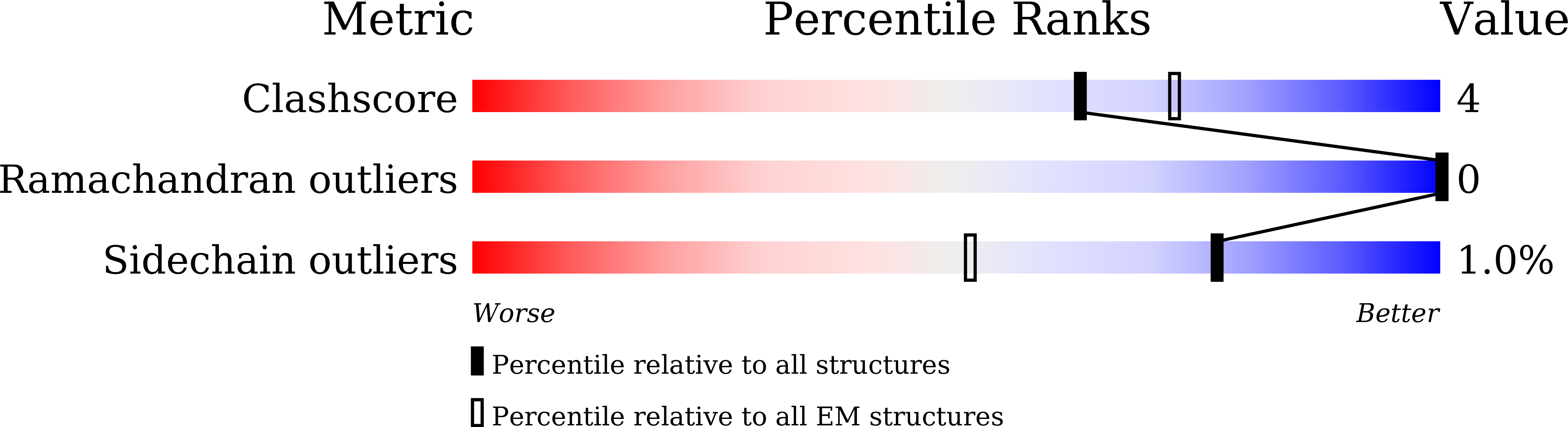Constitutive signal bias mediated by the human GHRHR splice variant 1.
Cong, Z., Zhou, F., Zhang, C., Zou, X., Zhang, H., Wang, Y., Zhou, Q., Cai, X., Liu, Q., Li, J., Shao, L., Mao, C., Wang, X., Wu, J., Xia, T., Zhao, L.H., Jiang, H., Zhang, Y., Xu, H.E., Cheng, X., Yang, D., Wang, M.W.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34599099
- DOI: https://doi.org/10.1073/pnas.2106606118
- Primary Citation of Related Structures:
7V9L, 7V9M - PubMed Abstract:
Alternative splicing of G protein-coupled receptors has been observed, but their functions are largely unknown. Here, we report that a splice variant (SV1) of the human growth hormone-releasing hormone receptor (GHRHR) is capable of transducing biased signal. Differing only at the receptor N terminus, GHRHR predominantly activates G s while SV1 selectively couples to β-arrestins. Based on the cryogenic electron microscopy structures of SV1 in the apo state or GHRH-bound state in complex with the G s protein, molecular dynamics simulations reveal that the N termini of GHRHR and SV1 differentiate the downstream signaling pathways, G s versus β-arrestins. As suggested by mutagenesis and functional studies, it appears that GHRH-elicited signal bias toward β-arrestin recruitment is constitutively mediated by SV1. The level of SV1 expression in prostate cancer cells is also positively correlated with ERK1/2 phosphorylation but negatively correlated with cAMP response. Our findings imply that constitutive signal bias may be a mechanism that ensures cancer cell proliferation.
Organizational Affiliation:
Department of Pharmacology, School of Basic Medical Sciences, Fudan University, Shanghai 200032, China.