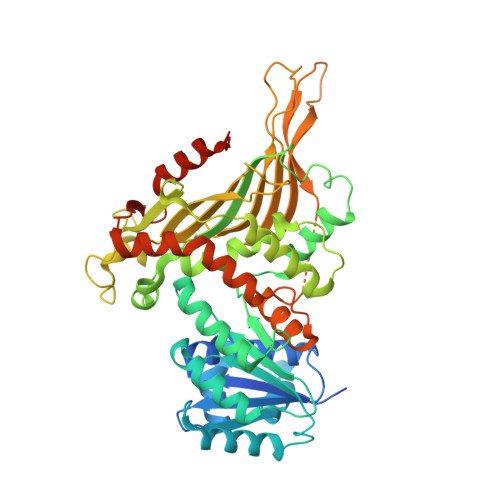
Allosteric role of a structural NADP + molecule in glucose-6-phosphate dehydrogenase activity.
Wei, X., Kixmoeller, K., Baltrusaitis, E., Yang, X., Marmorstein, R.(2022) Proc Natl Acad Sci U S A 119: e2119695119-e2119695119
- PubMed: 35858355
- DOI: https://doi.org/10.1073/pnas.2119695119
- Primary Citation of Related Structures:
7SNF, 7SNG, 7SNH, 7SNI, 7TOE, 7TOF, 7UAL, 7UC2 - PubMed Abstract:
Human glucose-6-phosphate dehydrogenase (G6PD) is the main cellular source of NADPH, and thus plays a key role in maintaining reduced glutathione to protect cells from oxidative stress disorders such as hemolytic anemia. G6PD is a multimeric enzyme that uses the cofactors β-D-glucose 6-phosphate (G6P) and "catalytic" NADP + (NADP + c), as well as a "structural" NADP + (NADP + s) located ∼25 Å from the active site, to generate NADPH. While X-ray crystallographic and biochemical studies have revealed a role for NADP + s in maintaining the catalytic activity by stabilizing the multimeric G6PD conformation, other potential roles for NADP + s have not been evaluated. Here, we determined the high resolution cryo-electron microscopy structures of human wild-type G6PD in the absence of bound ligands and a catalytic G6PD-D200N mutant bound to NADP + c and NADP + s in the absence or presence of G6P. A comparison of these structures, together with previously reported structures, reveals that the unliganded human G6PD forms a mixture of dimers and tetramers with similar overall folds, and binding of NADP + s induces a structural ordering of a C-terminal extension region and allosterically regulates G6P binding and catalysis. These studies have implications for understanding G6PD deficiencies and for therapy of G6PD-mediated disorders.
Organizational Affiliation:
Department of Biochemistry and Biophysics, University of Pennsylvania, Philadelphia, PA, 19104.