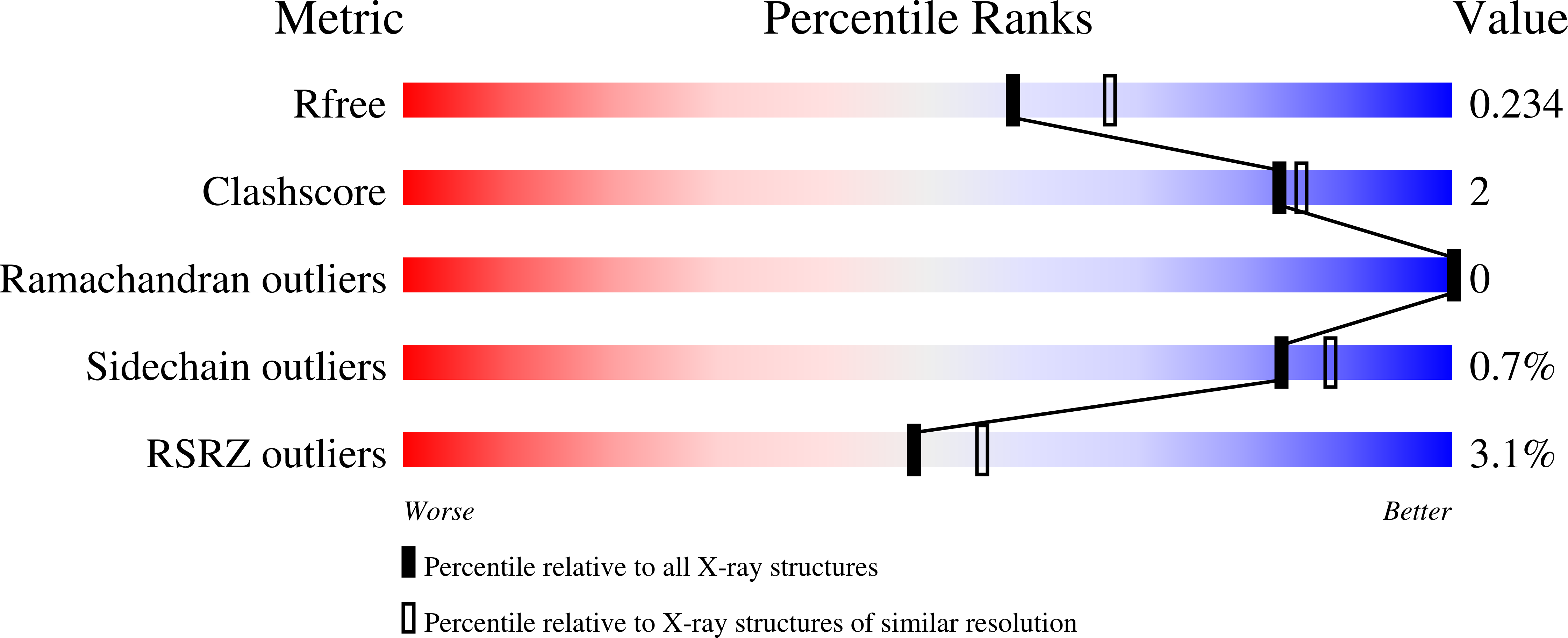
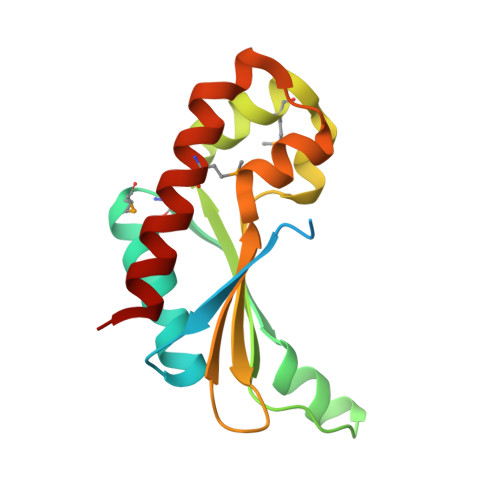
Crystal Structure of the Putative Fluoride Ion Transporter CrcB Bab1_1389 from Brucella abortus
Kim, Y., Tesar, C., Pastore, T., Endres, M., Babnigg, G., Crosson, S., Joachimiak, A., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.