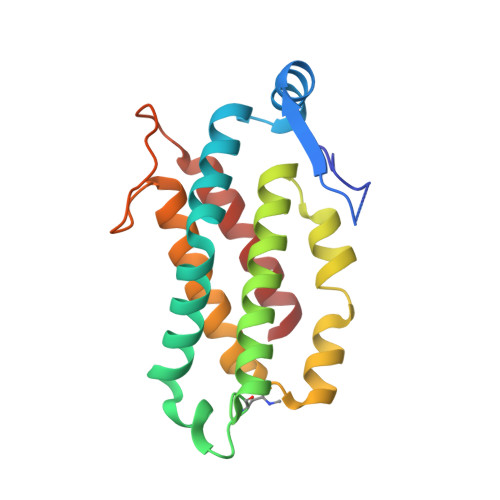
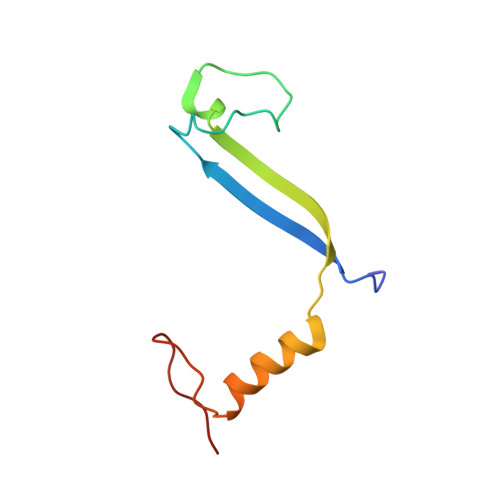
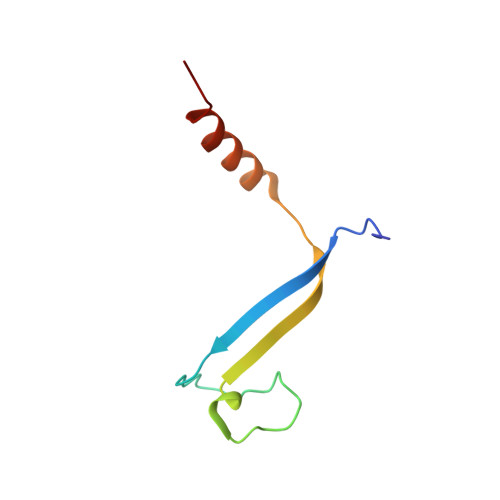
Molecular structures reveal the origin of spectral variation in cryptophyte light harvesting antenna proteins.
Michie, K.A., Harrop, S.J., Rathbone, H.W., Wilk, K.E., Teng, C.Y., Hoef-Emden, K., Hiller, R.G., Green, B.R., Curmi, P.M.G.(2023) Protein Sci 32: e4586-e4586
- PubMed: 36721353
- DOI: https://doi.org/10.1002/pro.4586
- Primary Citation of Related Structures:
7T7U, 7T89, 7T8S - PubMed Abstract:
In addition to their membrane-bound chlorophyll a/c light-harvesting antenna, the cryptophyte algae have evolved a unique phycobiliprotein antenna system located in the thylakoid lumen. The basic unit of this antenna consists of two copies of an αβ protomer where the α and β subunits scaffold different combinations of a limited number of linear tetrapyrrole chromophores. While the β subunit is highly conserved, encoded by a single plastid gene, the nuclear-encoded α subunits have evolved diversified multigene families. It is still unclear how this sequence diversity results in the spectral diversity of the mature proteins. By careful examination of three newly determined crystal structures in comparison with three previously obtained, we show how the α subunit amino acid sequences control chromophore conformations and hence spectral properties even when the chromophores are identical. Previously we have shown that α subunits control the quaternary structure of the mature αβ.αβ complex (either open or closed), however, each species appeared to only harbor a single quaternary form. Here we show that species of the Hemiselmis genus contain expressed α subunit genes that encode both distinct quaternary structures. Finally, we have discovered a common single-copy gene (expressed into protein) consisting of tandem copies of a small α subunit that could potentially scaffold pairs of light harvesting units. Together, our results show how the diversity of the multigene α subunit family produces a range of mature cryptophyte antenna proteins with differing spectral properties, and the potential for minor forms that could contribute to acclimation to varying light regimes.
Organizational Affiliation:
School of Physics, The University of New South Wales, Sydney, New South Wales, Australia.