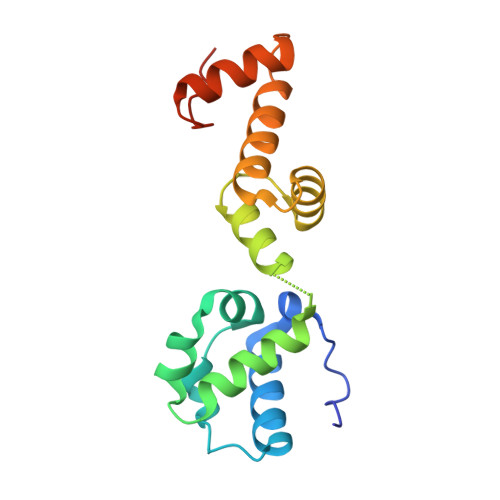
Fusion crystallization reveals the behavior of both the 1TEL crystallization chaperone and the TNK1 UBA domain.
Nawarathnage, S., Tseng, Y.J., Soleimani, S., Smith, T., Pedroza Romo, M.J., Abiodun, W.O., Egbert, C.M., Madhusanka, D., Bunn, D., Woods, B., Tsubaki, E., Stewart, C., Brown, S., Doukov, T., Andersen, J.L., Moody, J.D.(2023) Structure 31: 1589
- PubMed: 37776857
- DOI: https://doi.org/10.1016/j.str.2023.09.001
- Primary Citation of Related Structures:
7T8J, 7TCY, 7TDY, 7U4W, 7U4Z - PubMed Abstract:
Human thirty-eight-negative kinase-1 (TNK1) is implicated in cancer progression. The TNK1 ubiquitin-associated (UBA) domain binds polyubiquitin and plays a regulatory role in TNK1 activity and stability. No experimentally determined molecular structure of this unusual UBA domain is available. We fused the UBA domain to the 1TEL variant of the translocation ETS leukemia protein sterile alpha motif (TELSAM) crystallization chaperone and obtained crystals diffracting as far as 1.53 Å. GG and GSGG linkers allowed the UBA to reproducibly find a productive binding mode against its host 1TEL polymer and crystallize at protein concentrations as low as 0.2 mg/mL. Our studies support a mechanism of 1TEL fusion crystallization and show that 1TEL fusion crystals require fewer crystal contacts than traditional protein crystals. Modeling and experimental validation suggest the UBA domain may be selective for both the length and linkages of polyubiquitin chains.
Organizational Affiliation:
Department of Chemistry and Biochemistry, Brigham Young University, Provo, UT, USA.