Regulation of multiple dimeric states of E-cadherin by adhesion activating antibodies revealed through Cryo-EM and X-ray crystallography.
Maker, A., Bolejack, M., Schecterson, L., Hammerson, B., Abendroth, J., Edwards, T.E., Staker, B., Myler, P.J., Gumbiner, B.M.(2022) PNAS Nexus 1: pgac163-pgac163
- PubMed: 36157596
- DOI: https://doi.org/10.1093/pnasnexus/pgac163
- Primary Citation of Related Structures:
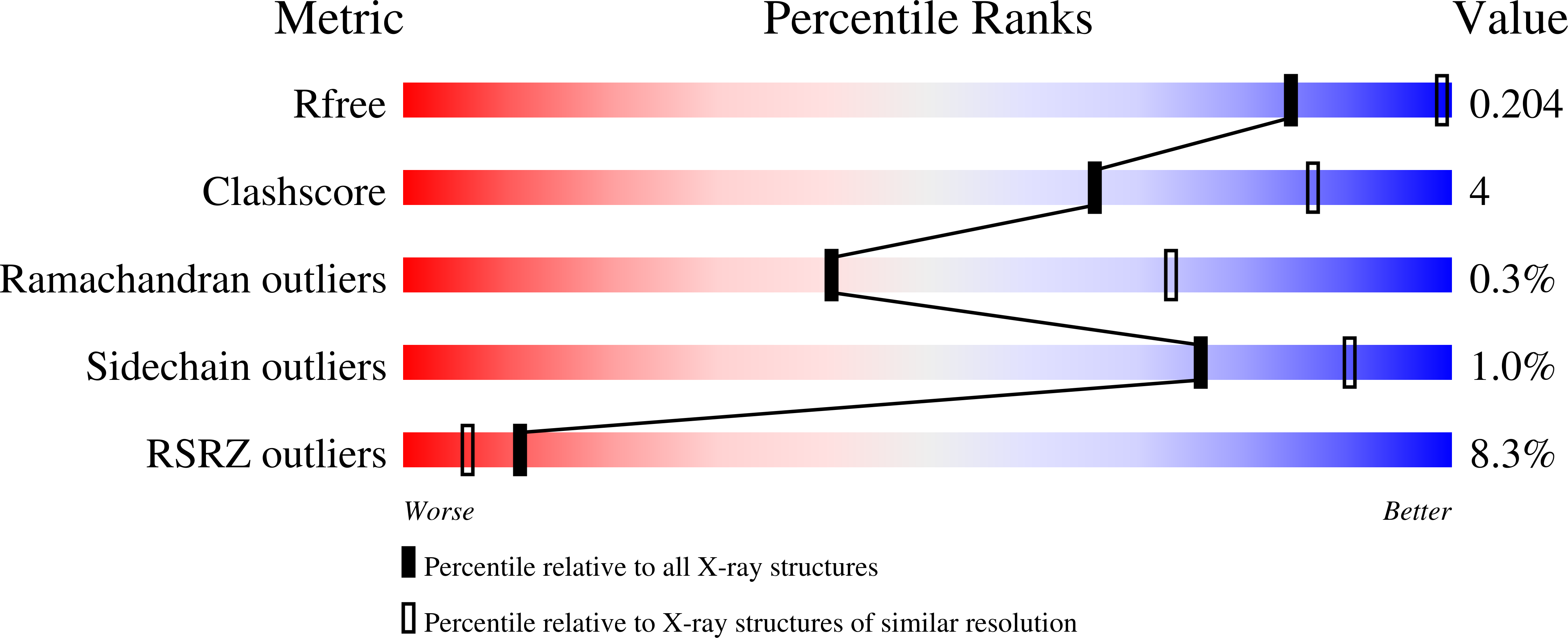
6VEL, 7STZ - PubMed Abstract:
E-cadherin adhesion is regulated at the cell surface, a process that can be replicated by activating antibodies. We use cryo-electron microscopy (EM) and X-ray crystallography to examine functional states of the cadherin adhesive dimer. This dimer is mediated by N-terminal beta strand-swapping involving Trp2, and forms via a different transient X-dimer intermediate. X-dimers are observed in cryo-EM along with monomers and strand-swap dimers, indicating that X-dimers form stable interactions. A novel EC4-mediated dimer was also observed. Activating Fab binding caused no gross structural changes in E-cadherin monomers, but can facilitate strand swapping. Moreover, activating Fab binding is incompatible with the formation of the X-dimer. Both cryo-EM and X-ray crystallography reveal a distinctive twisted strand-swap dimer conformation caused by an outward shift in the N-terminal beta strand that may represent a strengthened state. Thus, regulation of adhesion involves changes in cadherin dimer configurations.
Organizational Affiliation:
Department of Biochemistry, University of Washington, USA.