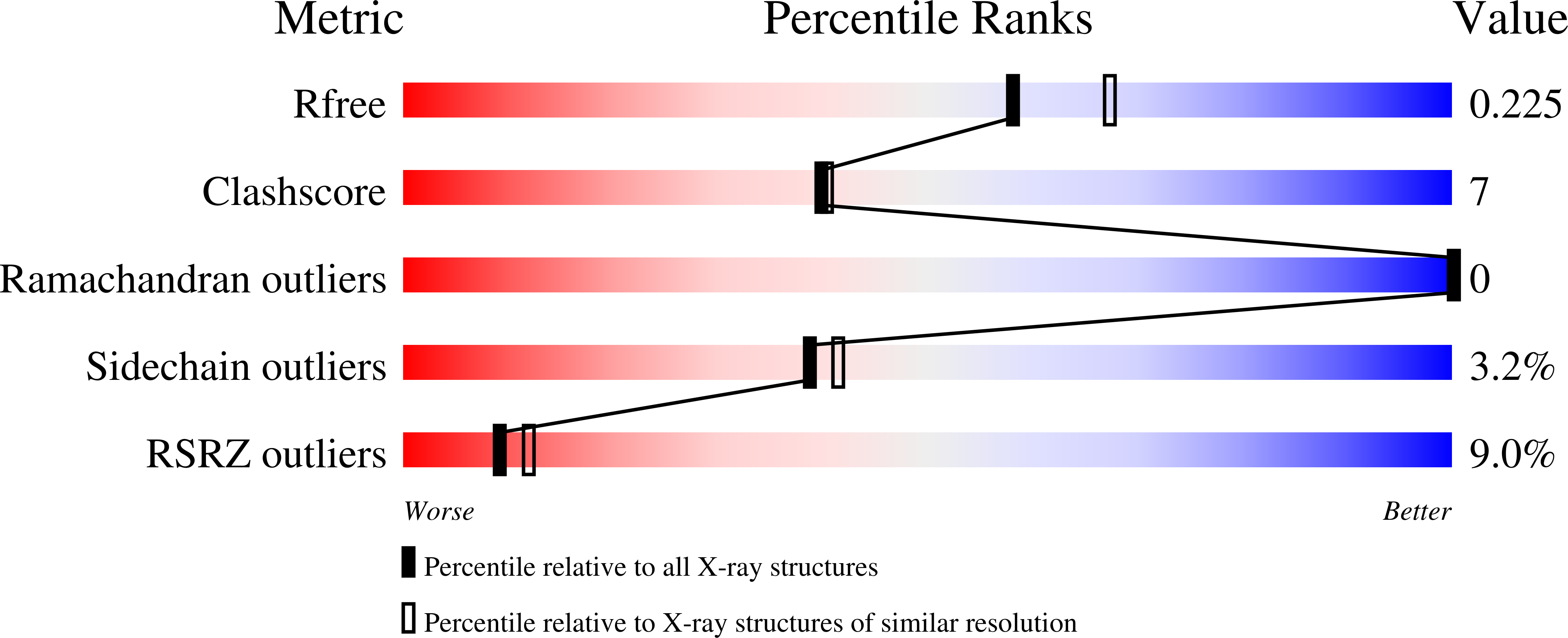
The active site region plays a critical role in Na + binding to thrombin.
Pelc, L.A., Koester, S.K., Kukla, C.R., Chen, Z., Di Cera, E.(2022) J Biol Chem 298: 101458-101458
- PubMed: 34861239
- DOI: https://doi.org/10.1016/j.jbc.2021.101458
- Primary Citation of Related Structures:
7SR9 - PubMed Abstract:
The catalytic activity of thrombin and other enzymes of the blood coagulation and complement cascades is enhanced significantly by binding of Na + to a site >15 Å away from the catalytic residue S195, buried within the 180 and 220 loops that also contribute to the primary specificity of the enzyme. Rapid kinetics support a binding mechanism of conformational selection where the Na + -binding site is in equilibrium between open (N) and closed (N ∗ ) forms and the cation binds selectively to the N form. Allosteric transduction of this binding step produces enhanced catalytic activity. Molecular details on how Na + gains access to this site and communicates allosterically with the active site remain poorly defined. In this study, we show that the rate of the N ∗ →N transition is strongly correlated with the analogous E ∗ →E transition that governs the interaction of synthetic and physiologic substrates with the active site. This correlation supports the active site as the likely point of entry for Na + to its binding site. Mutagenesis and structural data rule out an alternative path through the pore defined by the 180 and 220 loops. We suggest that the active site communicates allosterically with the Na + site through a network of H-bonded water molecules that embeds the primary specificity pocket. Perturbation of the mobility of S195 and its H-bonding capabilities alters interaction with this network and influences the kinetics of Na + binding and allosteric transduction. These findings have general mechanistic relevance for Na + -activated proteases and allosteric enzymes.
Organizational Affiliation:
Edward A. Doisy Department of Biochemistry and Molecular Biology, Saint Louis University School of Medicine, St. Louis, Missouri, USA.