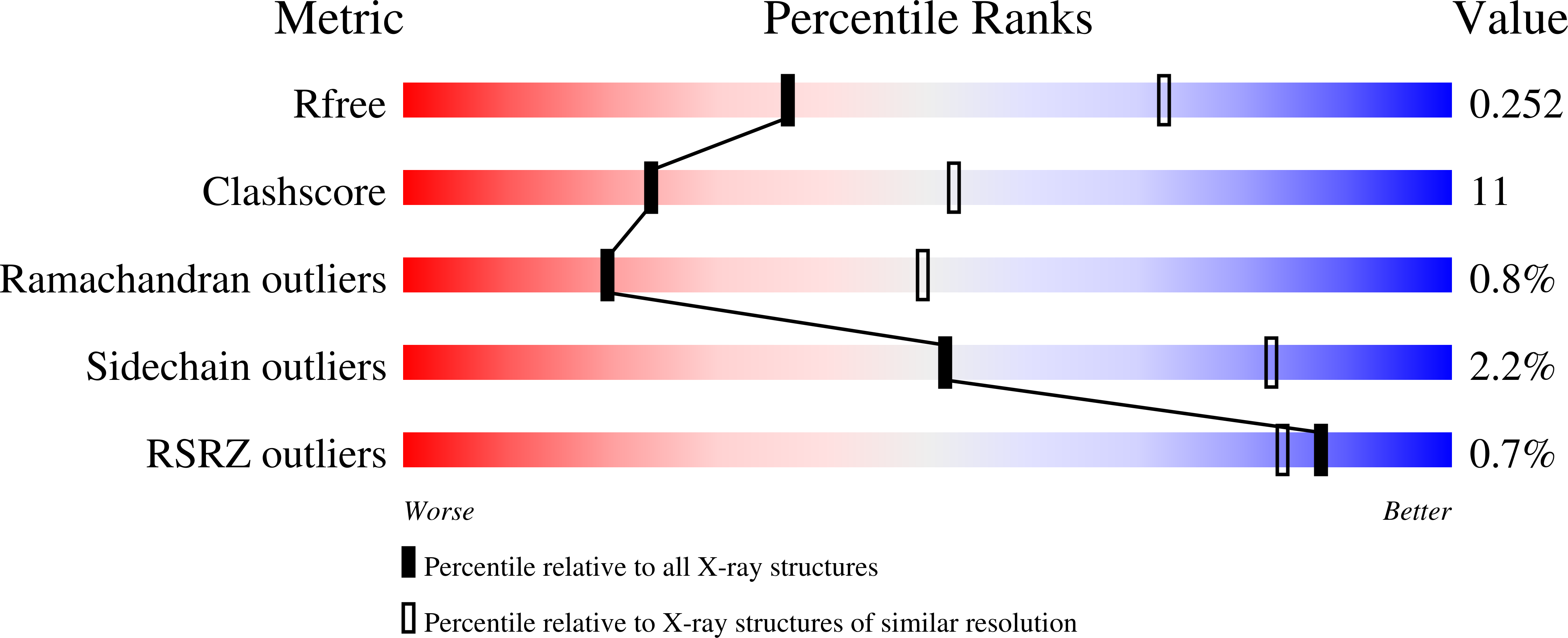
Effects of the T337M and G391V disease-related variants on human phosphoglucomutase 1: structural disruptions large and small.
Stiers, K.M., Owuocha, L.F., Beamer, L.J.(2022) Acta Crystallogr F Struct Biol Commun 78: 200-209
- PubMed: 35506765
- DOI: https://doi.org/10.1107/S2053230X22004174
- Primary Citation of Related Structures:
7S0W, 7S77 - PubMed Abstract:
Phosphoglucomutase 1 (PGM1) plays a central role in glucose homeostasis in human cells. Missense variants of this enzyme cause an inborn error of metabolism, which is categorized as a congenital disorder of glycosylation. Here, two disease-related variants of PGM1, T337M and G391V, which are both located in domain 3 of the four-domain protein, were characterized via X-ray crystallography and biochemical assays. The studies show multiple impacts resulting from these dysfunctional variants, including both short- and long-range structural perturbations. In the T337M variant these are limited to a small shift in an active-site loop, consistent with reduced enzyme activity. In contrast, the G391V variant produces a cascade of structural perturbations, including displacement of both the catalytic phosphoserine and metal-binding loops. This work reinforces several themes that were found in prior studies of dysfunctional PGM1 variants, including increased structural flexibility and the outsized impacts of mutations affecting interdomain interfaces. The molecular mechanisms of PGM1 variants have implications for newly described inherited disorders of related enzymes.
Organizational Affiliation:
Biochemistry Department, University of Missouri, Columbia, MO 65211, USA.