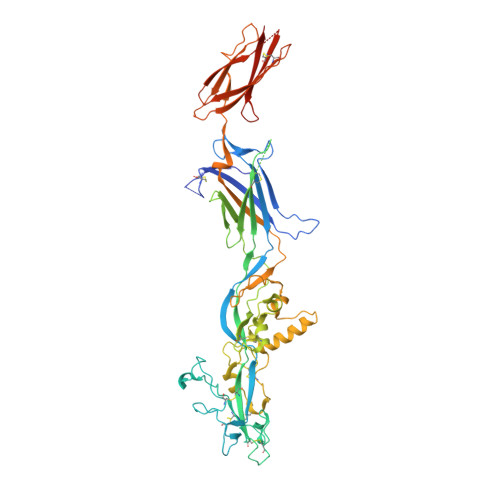
Monomeric prefusion structure of an extremophile gamete fusogen and stepwise formation of the postfusion trimeric state.
Feng, J., Dong, X., Su, Y., Lu, C., Springer, T.A.(2022) Nat Commun 13: 4064-4064
- PubMed: 35831325
- DOI: https://doi.org/10.1038/s41467-022-31744-z
- Primary Citation of Related Structures:
7S0K - PubMed Abstract:
Here, we study the gamete fusogen HAP2 from Cyanidioschyzon merolae (Cyani), an extremophile red algae that grows at acidic pH at 45 °C. HAP2 has a trimeric postfusion structure with similarity to viral class II fusion proteins, but its prefusion structure has been elusive. The crystal structure of a monomeric prefusion state of Cyani HAP2 shows it is highly extended with three domains in the order D2, D1, and D3. Three hydrophobic fusion loops at the tip of D2 are each required for postfusion state formation. We followed by negative stain electron microscopy steps in the process of detergent micelle-stimulated postfusion state formation. In an intermediate state, two or three linear HAP2 monomers associate at the end of D2 bearing its fusion loops. Subsequently, D2 and D1 line the core of a trimer and D3 folds back over the exterior of D1 and D2. D3 is not required for formation of intermediate or postfusion-like states.
Organizational Affiliation:
Program in Cellular and Molecular Medicine, Department of Pediatrics, Boston Children's Hospital, Boston, MA, USA.