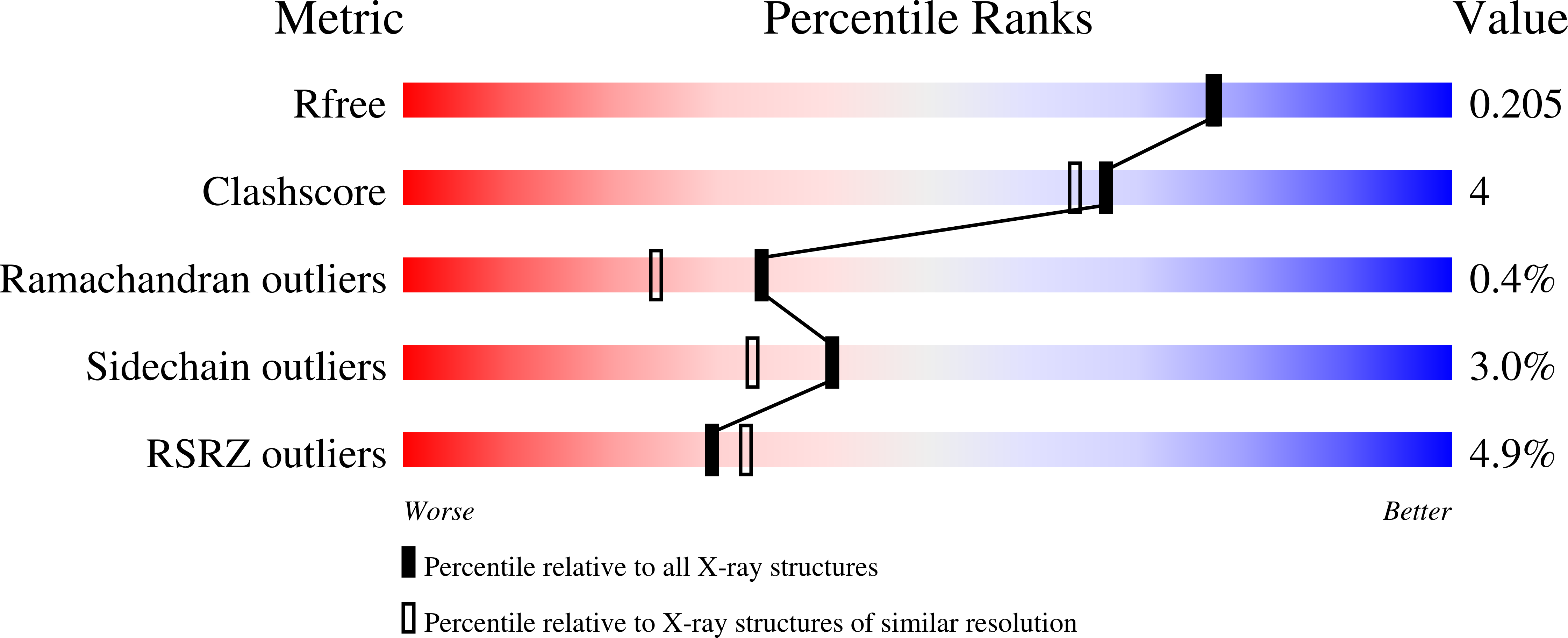
Conformational flexibility in carbapenem hydrolysis drives substrate specificity of the class D carbapenemase OXA-24/40.
Mitchell, J.M., June, C.M., Baggett, V.L., Lowe, B.C., Ruble, J.F., Bonomo, R.A., Leonard, D.A., Powers, R.A.(2022) J Biol Chem 298: 102127-102127
- PubMed: 35709986
- DOI: https://doi.org/10.1016/j.jbc.2022.102127
- Primary Citation of Related Structures:
7RP8, 7RP9, 7RPA, 7RPB, 7RPC, 7RPD, 7RPE, 7RPF, 7RPG - PubMed Abstract:
The evolution of multidrug resistance in Acinetobacter spp. increases the risk of our best antibiotics losing their efficacy. From a clinical perspective, the carbapenem-hydrolyzing class D β-lactamase subfamily present in Acinetobacter spp. is particularly concerning because of its ability to confer resistance to carbapenems. The kinetic profiles of class D β-lactamases exhibit variability in carbapenem hydrolysis, suggesting functional differences. To better understand the structure-function relationship between the carbapenem-hydrolyzing class D β-lactamase OXA-24/40 found in Acinetobacter baumannii and carbapenem substrates, we analyzed steady-state kinetics with the carbapenem antibiotics meropenem and ertapenem and determined the structures of complexes of OXA-24/40 bound to imipenem, meropenem, doripenem, and ertapenem, as well as the expanded-spectrum cephalosporin cefotaxime, using X-ray crystallography. We show that OXA-24/40 exhibits a preference for ertapenem compared with meropenem, imipenem, and doripenem, with an increase in catalytic efficiency of up to fourfold. We suggest that superposition of the nine OXA-24/40 complexes will better inform future inhibitor design efforts by providing insight into the complicated and varying ways in which carbapenems are selected and bound by class D β-lactamases.
Organizational Affiliation:
Department of Chemistry, Grand Valley State University, Allendale, Michigan, USA.