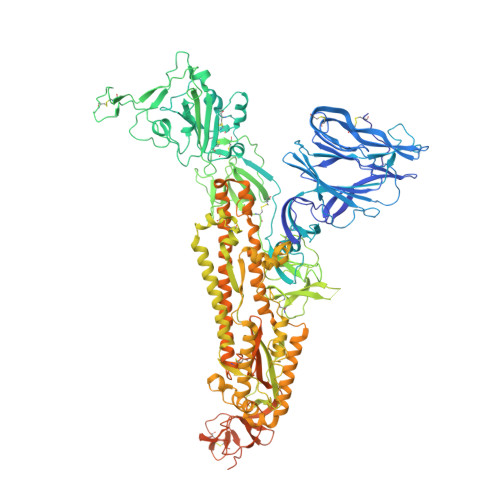
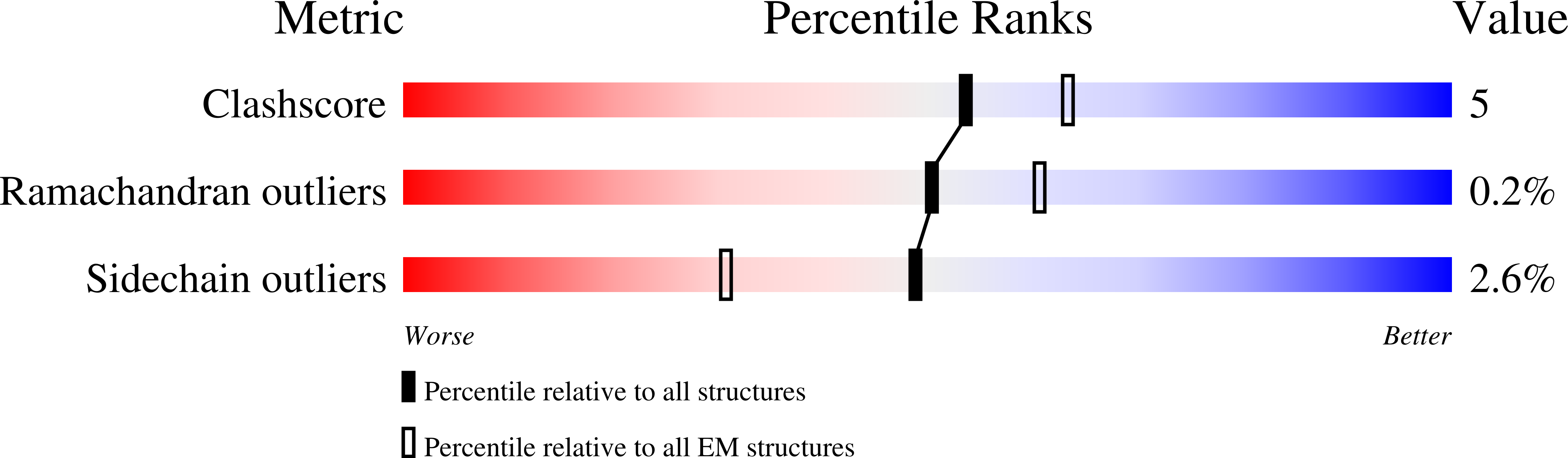Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Buchanan, C.J., Gaunt, B., Harrison, P.J., Yang, Y., Liu, J., Khan, A., Giltrap, A.M., Le Bas, A., Ward, P.N., Gupta, K., Dumoux, M., Tan, T.K., Schimaski, L., Daga, S., Picchiotti, N., Baldassarri, M., Benetti, E., Fallerini, C., Fava, F., Giliberti, A., Koukos, P.I., Davy, M.J., Lakshminarayanan, A., Xue, X., Papadakis, G., Deimel, L.P., Casablancas-Antras, V., Claridge, T.D.W., Bonvin, A.M.J.J., Sattentau, Q.J., Furini, S., Gori, M., Huo, J., Owens, R.J., Schaffitzel, C., Berger, I., Renieri, A., Naismith, J.H., Baldwin, A.J., Davis, B.G.(2022) Science 377: eabm3125-eabm3125
- PubMed: 35737812
- DOI: https://doi.org/10.1126/science.abm3125
- Primary Citation of Related Structures:
7QUR, 7QUS - PubMed Abstract:
Many pathogens exploit host cell-surface glycans. However, precise analyses of glycan ligands binding with heavily modified pathogen proteins can be confounded by overlapping sugar signals and/or compounded with known experimental constraints. Universal saturation transfer analysis (uSTA) builds on existing nuclear magnetic resonance spectroscopy to provide an automated workflow for quantitating protein-ligand interactions. uSTA reveals that early-pandemic, B-origin-lineage severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike trimer binds sialoside sugars in an "end-on" manner. uSTA-guided modeling and a high-resolution cryo-electron microscopy structure implicate the spike N-terminal domain (NTD) and confirm end-on binding. This finding rationalizes the effect of NTD mutations that abolish sugar binding in SARS-CoV-2 variants of concern. Together with genetic variance analyses in early pandemic patient cohorts, this binding implicates a sialylated polylactosamine motif found on tetraantennary N-linked glycoproteins deep in the human lung as potentially relevant to virulence and/or zoonosis.
Organizational Affiliation:
Rosalind Franklin Institute, Harwell Science and Innovation Campus, Oxford OX11 0FA, UK.