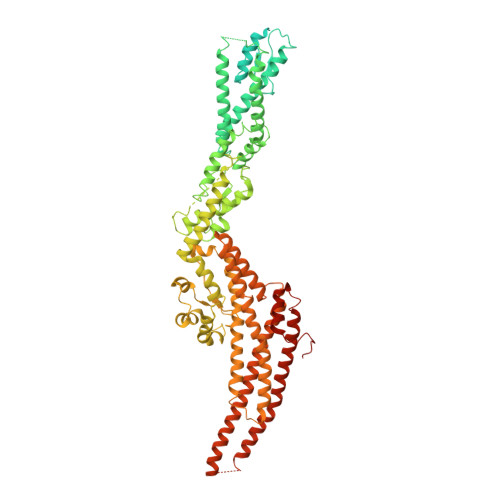
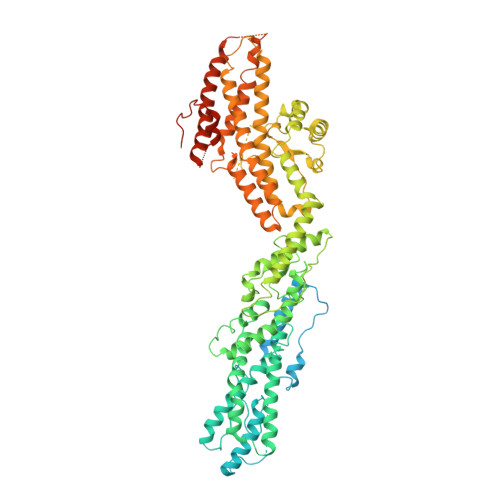
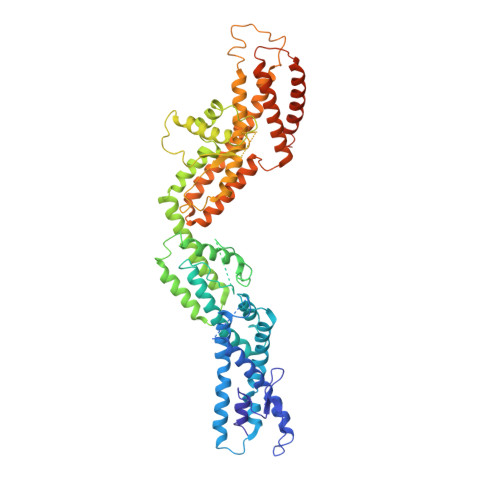
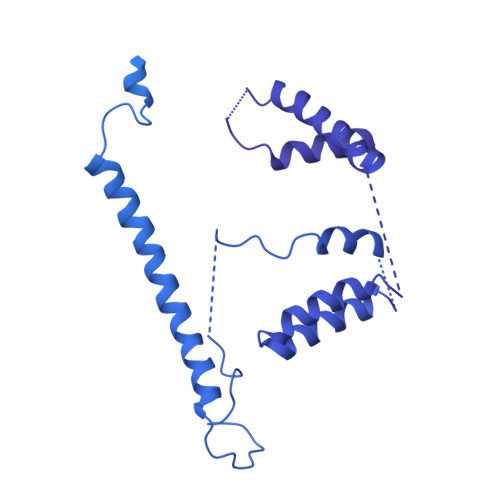
Modular assembly of the principal microtubule nucleator gamma-TuRC.
Wurtz, M., Zupa, E., Atorino, E.S., Neuner, A., Bohler, A., Rahadian, A.S., Vermeulen, B.J.A., Tonon, G., Eustermann, S., Schiebel, E., Pfeffer, S.(2022) Nat Commun 13: 473-473
- PubMed: 35078983
- DOI: https://doi.org/10.1038/s41467-022-28079-0
- Primary Citation of Related Structures:
7QJ0, 7QJ1, 7QJ2, 7QJ3, 7QJ4, 7QJ5, 7QJ6, 7QJ7, 7QJ8, 7QJ9, 7QJA, 7QJB, 7QJC, 7QJD, 7QJE - PubMed Abstract:
The gamma-tubulin ring complex (γ-TuRC) is the principal microtubule nucleation template in vertebrates. Recent cryo-EM reconstructions visualized the intricate quaternary structure of the γ-TuRC, containing more than thirty subunits, raising fundamental questions about γ-TuRC assembly and the role of actin as an integral part of the complex. Here, we reveal the structural mechanism underlying modular γ-TuRC assembly and identify a functional role of actin in microtubule nucleation. During γ-TuRC assembly, a GCP6-stabilized core comprising GCP2-3-4-5-4-6 is expanded by stepwise recruitment, selective stabilization and conformational locking of four pre-formed GCP2-GCP3 units. Formation of the lumenal bridge specifies incorporation of the terminal GCP2-GCP3 unit and thereby leads to closure of the γ-TuRC ring in a left-handed spiral configuration. Actin incorporation into the complex is not relevant for γ-TuRC assembly and structural integrity, but determines γ-TuRC geometry and is required for efficient microtubule nucleation and mitotic chromosome alignment in vivo.
Organizational Affiliation:
Zentrum für Molekulare Biologie der Universität Heidelberg, DKFZ-ZMBH Allianz, Im Neuenheimer Feld 282, 69120, Heidelberg, Germany.