The mechanism of allosteric activation of SYK kinase derived from multiple phospho-ITAM-bound structures
Bradshaw, W.J., Harris, G., Gileadi, O., Katis, V.L.(2024) Structure
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(2024) Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tyrosine-protein kinase SYK | 265 | Homo sapiens | Mutation(s): 0 Gene Names: SYK EC: 2.7.10.2 | 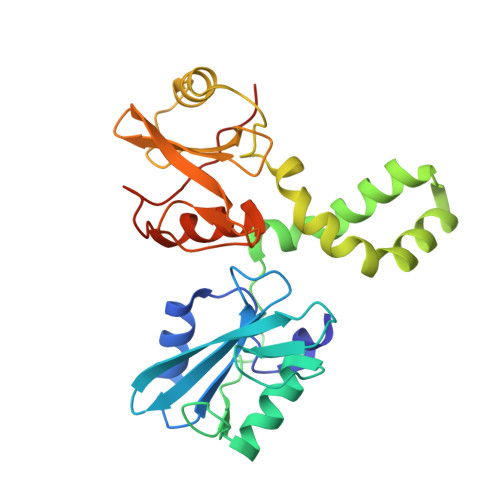 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P43405 (Homo sapiens) Explore P43405 Go to UniProtKB: P43405 | |||||
PHAROS: P43405 GTEx: ENSG00000165025 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P43405 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| High affinity immunoglobulin epsilon receptor subunit gamma | 20 | Homo sapiens | Mutation(s): 0 | 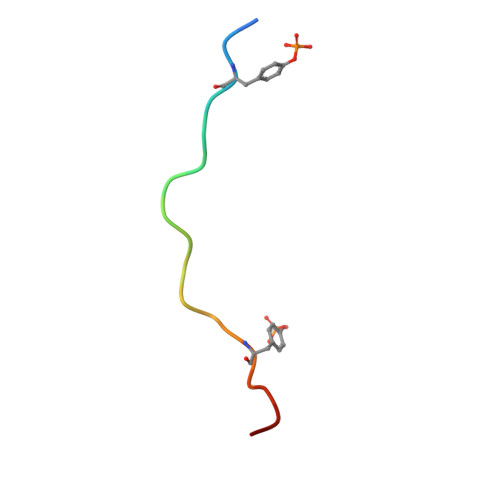 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P30273 (Homo sapiens) Explore P30273 Go to UniProtKB: P30273 | |||||
PHAROS: P30273 GTEx: ENSG00000158869 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P30273 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PEG Query on PEG | R [auth BBB], Z [auth EEE] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| PO4 Query on PO4 | M [auth AAA] N [auth AAA] P [auth BBB] Q [auth BBB] S [auth CCC] | PHOSPHATE ION O4 P NBIIXXVUZAFLBC-UHFFFAOYSA-K |  | ||
| EDO Query on EDO | O [auth AAA], U [auth CCC], V [auth CCC] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| PTR Query on PTR | G [auth GGG] H [auth HHH] I [auth III] J [auth JJJ] K [auth KKK] G [auth GGG], H [auth HHH], I [auth III], J [auth JJJ], K [auth KKK], L [auth LLL] | L-PEPTIDE LINKING | C9 H12 N O6 P |  | TYR |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 144.136 | α = 90 |
| b = 88.035 | β = 113.575 |
| c = 158.486 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| DIALS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute on Aging (NIH/NIA) | United States | 5U54AG065187-03 |