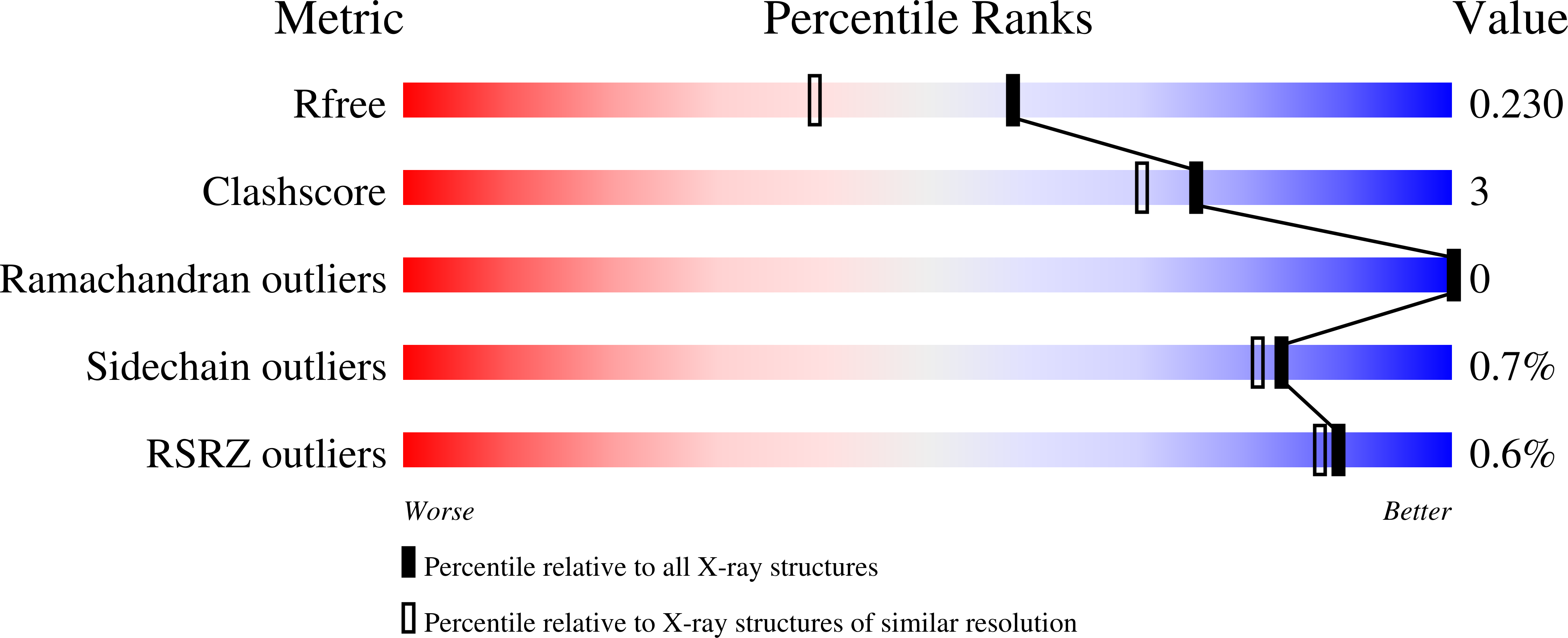
New ligand-binding sites identified in the crystal structures of [beta]-lactoglobulin complexes with desipramine
Loch, J.I., Barciszewski, J., Sliwiak, J., Bonarek, P., Wrobel, P., Pokrywka, K., Shabalin, I.G., Minor, W., Jaskolski, M., Lewinski, K.(2022) IUCrJ 9: 386-398