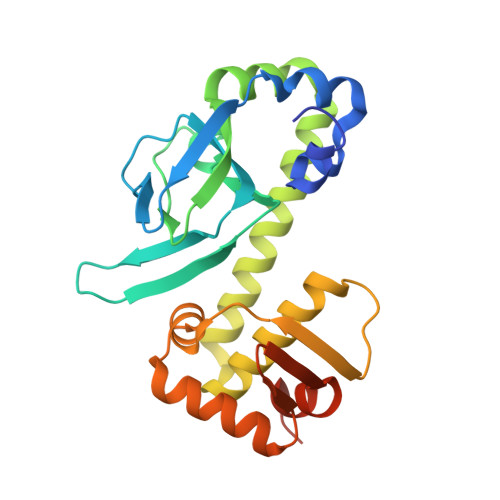
Structural Basis of Dual Specificity of Sinorhizobium meliloti Clr, a cAMP and cGMP Receptor Protein.
Werel, L., Farmani, N., Krol, E., Serrania, J., Essen, L.O., Becker, A.(2023) mBio 14: e0302822-e0302822
- PubMed: 37017526
- DOI: https://doi.org/10.1128/mbio.03028-22
- Primary Citation of Related Structures:
7PZA, 7PZB - PubMed Abstract:
In bacteria, the most prevalent receptor proteins of 3',5'-cyclic AMP (cAMP) and 3',5'-cyclic GMP (cGMP) are found among transcription factors of the Crp-Fnr superfamily. The prototypic Escherichia coli catabolite activator protein (CAP) represents the main Crp cluster of this superfamily and is known to bind cAMP and cGMP but to mediate transcription activation only in its cAMP-bound state. In contrast, both cyclic nucleotides mediate transcription activation by Sinorhizobium meliloti Clr, mapping to cluster G of Crp-like proteins. We present crystal structures of Clr-cAMP and Clr-cGMP bound to the core motif of the palindromic Clr DNA binding site (CBS). We show that both cyclic nucleotides shift ternary Clr-cNMP-CBS-DNA complexes (where cNMP is cyclic nucleotide monophosphate) to almost identical active conformations, unlike the situation known for the E. coli CAP-cNMP complex. Isothermal titration calorimetry measured similar affinities of cAMP and cGMP binding to Clr in the presence of CBS core motif DNA (equilibrium dissociation constant for cNMP ( K D cNMP ], ~7 to 11 μM). However, different affinities were determined in the absence of this DNA ( K D cGMP , ~24 μM; K D cAMP , ~6 μM). Sequencing of Clr-coimmunoprecipitated DNA as well as electrophoretic mobility shift and promoter-probe assays expanded the list of experimentally proven Clr-regulated promoters and CBS. This comprehensive set of CBS features conserved nucleobases that are consistent with the sequence readout through interactions of Clr amino acid residues with these nucleobases, as revealed by the Clr-cNMP-CBS-DNA crystal structures. IMPORTANCE Cyclic 3',5'-AMP (cAMP) and cyclic 3',5'-GMP (cGMP) are both long known as important nucleotide secondary messengers in eukaryotes. This is also the case for cAMP in prokaryotes, whereas a signaling role for cGMP in this domain of life has been recognized only recently. Catabolite repressor proteins (CRPs) are the most ubiquitous bacterial cAMP receptor proteins. Escherichia coli CAP, the prototypic transcription regulator of the main Crp cluster, binds both cyclic mononucleotides, but only the CAP-cAMP complex promotes transcription activation. In contrast, Crp cluster G proteins studied so far are activated by cGMP or by both cAMP and cGMP. Here, we report a structural analysis of the cAMP- and cGMP-activatable cluster G member Clr from Sinorhizobium meliloti, how binding of cAMP and cGMP shifts Clr to its active conformation, and the structural basis of its DNA binding site specificity.
Organizational Affiliation:
Department of Chemistry, Philipps-Universität Marburg, Marburg, Germany.