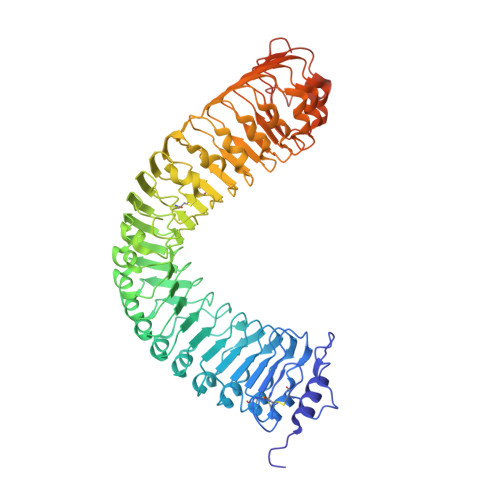
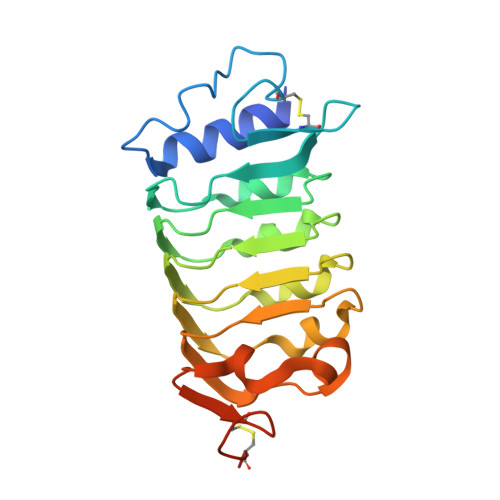
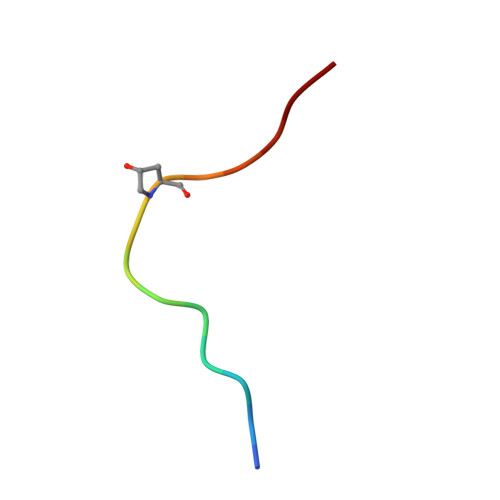
HSL1 and BAM1/2 impact epidermal cell development by sensing distinct signaling peptides.
Roman, A.O., Jimenez-Sandoval, P., Augustin, S., Broyart, C., Hothorn, L.A., Santiago, J.(2022) Nat Commun 13: 876-876
- PubMed: 35169143
- DOI: https://doi.org/10.1038/s41467-022-28558-4
- Primary Citation of Related Structures:
7ODK, 7ODV, 7OGO, 7OGQ, 7OGU, 7OGZ - PubMed Abstract:
The membrane receptor kinases HAESA and HSL2 recognize a family of IDA/IDL signaling peptides to control cell separation processes in different plant organs. The homologous HSL1 has been reported to regulate epidermal cell patterning by interacting with a different class of signaling peptides from the CLE family. Here we demonstrate that HSL1 binds IDA/IDL peptides with high, and CLE peptides with lower affinity, respectively. Ligand sensing capability and receptor activation of HSL1 require a SERK co-receptor kinase. Crystal structures with IDA/IDLs or with CLE9 reveal that HSL1-SERK1 complex recognizes the entire IDA/IDL signaling peptide, while only parts of CLE9 are bound to the receptor. In contrast, the receptor kinase BAM1 interacts with the entire CLE9 peptide with high affinity and specificity. Furthermore, the receptor tandem BAM1/BAM2 regulates epidermal cell division homeostasis. Consequently, HSL1-IDLs and BAM1/BAM2-CLEs independently regulate cell patterning in the leaf epidermal tissue.
Organizational Affiliation:
The Plant Signaling Mechanisms Laboratory, Department of Plant Molecular Biology, University of Lausanne, 1015, Lausanne, Switzerland.