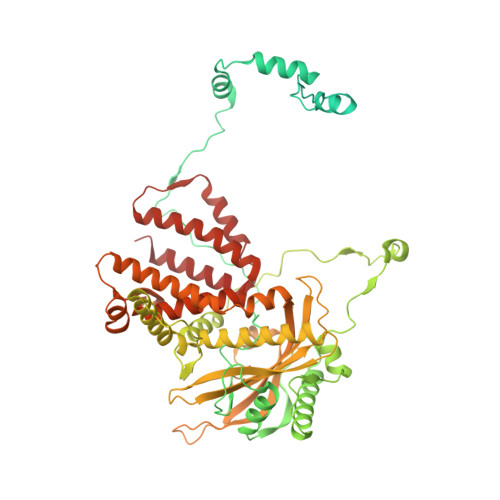
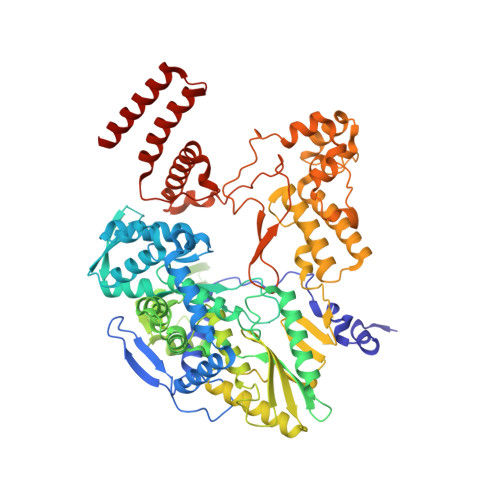
Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Keown, J.R., Zhu, Z., Carrique, L., Fan, H., Walker, A.P., Serna Martin, I., Pardon, E., Steyaert, J., Fodor, E., Grimes, J.M.(2022) Nat Commun 13: 251-251
- PubMed: 35017564
- DOI: https://doi.org/10.1038/s41467-021-27950-w
- Primary Citation of Related Structures:
7NFQ, 7NFR, 7NFT, 7NHA, 7NHC, 7NHX, 7NI0, 7NIK, 7NIL, 7NIR, 7NIS, 7NJ3, 7NJ4, 7NJ5, 7NJ7, 7NK1, 7NK2, 7NK4, 7NK6, 7NK8, 7NKA, 7NKC, 7NKI, 7NKR - PubMed Abstract:
Influenza A viruses cause seasonal epidemics and global pandemics, representing a considerable burden to healthcare systems. Central to the replication cycle of influenza viruses is the viral RNA-dependent RNA polymerase which transcribes and replicates the viral RNA genome. The polymerase undergoes conformational rearrangements and interacts with viral and host proteins to perform these functions. Here we determine the structure of the 1918 influenza virus polymerase in transcriptase and replicase conformations using cryo-electron microscopy (cryo-EM). We then structurally and functionally characterise the binding of single-domain nanobodies to the polymerase of the 1918 pandemic influenza virus. Combining these functional and structural data we identify five sites on the polymerase which are sensitive to inhibition by nanobodies. We propose that the binding of nanobodies at these sites either prevents the polymerase from assuming particular functional conformations or interactions with viral or host factors. The polymerase is highly conserved across the influenza A subtypes, suggesting these sites as effective targets for potential influenza antiviral development.
Organizational Affiliation:
Division of Structural Biology, Wellcome Centre for Human Genetics, University of Oxford, Oxford, UK.