Crystal Structure of chromodomain of CDYL in complex with inhibitor UNC6261
Beldar, S., Dong, A., Loppnau, P., Min, J., Arrowsmith, C.H., Edwards, A.M., Structural Genomics Consortium (SGC)To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Isoform 2 of Chromodomain Y-like protein | 57 | Homo sapiens | Mutation(s): 0 Gene Names: CDYL, CDYL1 EC: 4.2.1 | 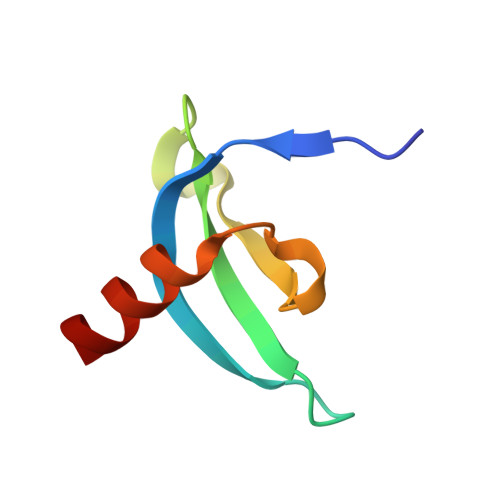 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9Y232 (Homo sapiens) Explore Q9Y232 Go to UniProtKB: Q9Y232 | |||||
PHAROS: Q9Y232 GTEx: ENSG00000153046 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9Y232 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| inhibitor UNC6261 | 6 | synthetic construct | Mutation(s): 0 | 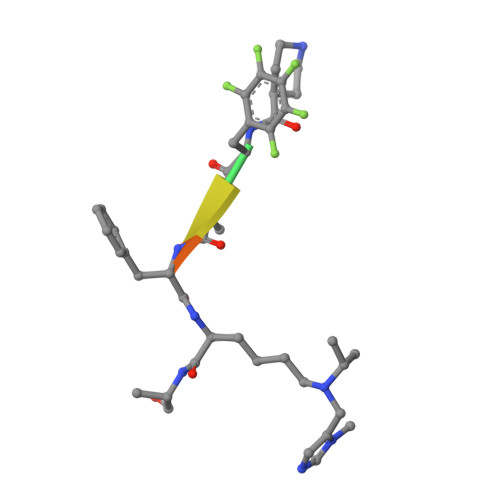 | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NA Query on NA | M [auth A], N [auth D] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| UNX Query on UNX | O [auth D], P [auth F] | UNKNOWN ATOM OR ION X |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| PF5 Query on PF5 | B [auth G] D [auth H] F [auth I] H [auth J] J [auth K] B [auth G], D [auth H], F [auth I], H [auth J], J [auth K], L | L-PEPTIDE LINKING | C9 H6 F5 N O2 |  | PHE |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 62.971 | α = 90 |
| b = 76.386 | β = 90 |
| c = 80.628 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-3000 | data scaling |
| PDB_EXTRACT | data extraction |
| PHASER | phasing |
| HKL-3000 | data reduction |