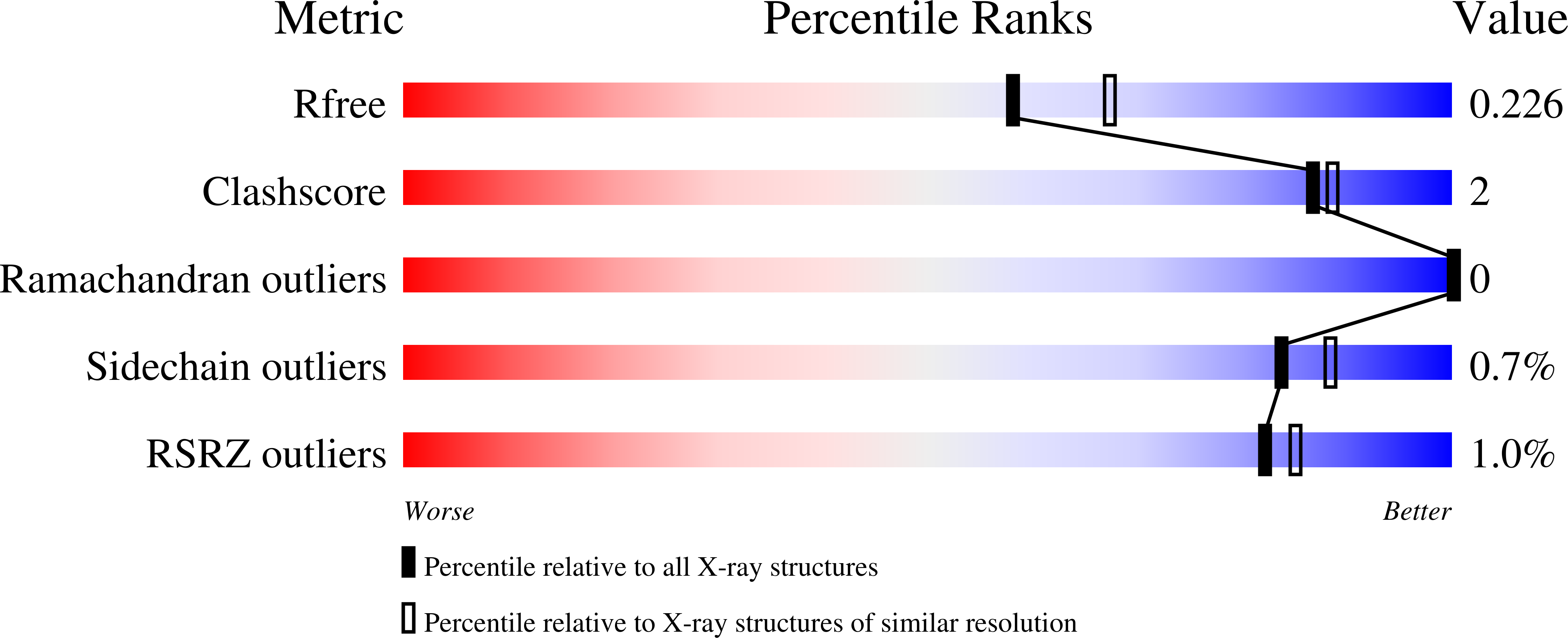
Efficacy of epetraborole against Mycobacterium abscessus is increased with norvaline.
Sullivan, J.R., Lupien, A., Kalthoff, E., Hamela, C., Taylor, L., Munro, K.A., Schmeing, T.M., Kremer, L., Behr, M.A.(2021) PLoS Pathog 17: e1009965-e1009965
- PubMed: 34637487
- DOI: https://doi.org/10.1371/journal.ppat.1009965
- Primary Citation of Related Structures:
7N11, 7N12 - PubMed Abstract:
Mycobacterium abscessus is the most common rapidly growing non-tuberculous mycobacteria to cause pulmonary disease in patients with impaired lung function such as cystic fibrosis. M. abscessus displays high intrinsic resistance to common antibiotics and inducible resistance to macrolides like clarithromycin. As such, M. abscessus is clinically resistant to the entire regimen of front-line M. tuberculosis drugs, and treatment with antibiotics that do inhibit M. abscessus in the lab results in cure rates of 50% or less. Here, we identified epetraborole (EPT) from the MMV pandemic response box as an inhibitor against the essential protein leucyl-tRNA synthetase (LeuRS) in M. abscessus. EPT protected zebrafish from lethal M. abscessus infection and did not induce self-resistance nor against clarithromycin. Contrary to most antimycobacterials, the whole-cell activity of EPT was greater against M. abscessus than M. tuberculosis, but crystallographic and equilibrium binding data showed that EPT binds LeuRSMabs and LeuRSMtb with similar residues and dissociation constants. Since EPT-resistant M. abscessus mutants lost LeuRS editing activity, these mutants became susceptible to misaminoacylation with leucine mimics like the non-proteinogenic amino acid norvaline. Proteomic analysis revealed that when M. abscessus LeuRS mutants were fed norvaline, leucine residues in proteins were replaced by norvaline, inducing the unfolded protein response with temporal changes in expression of GroEL chaperonins and Clp proteases. This supports our in vitro data that supplementation of media with norvaline reduced the emergence of EPT mutants in both M. abscessus and M. tuberculosis. Furthermore, the combination of EPT and norvaline had improved in vivo efficacy compared to EPT in a murine model of M. abscessus infection. Our results emphasize the effectiveness of EPT against the clinically relevant cystic fibrosis pathogen M. abscessus, and these findings also suggest norvaline adjunct therapy with EPT could be beneficial for M. abscessus and other mycobacterial infections like tuberculosis.
Organizational Affiliation:
Department of Microbiology & Immunology, McGill University, Montréal, Canada.