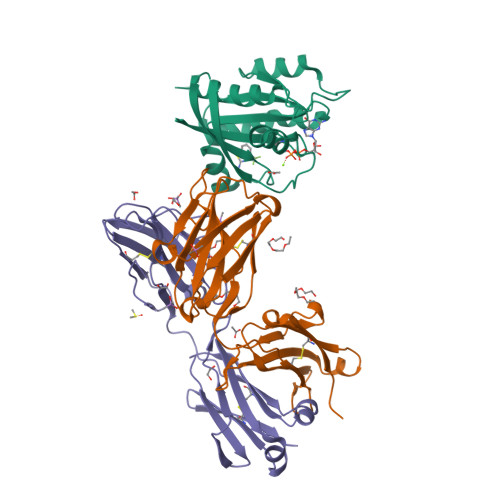
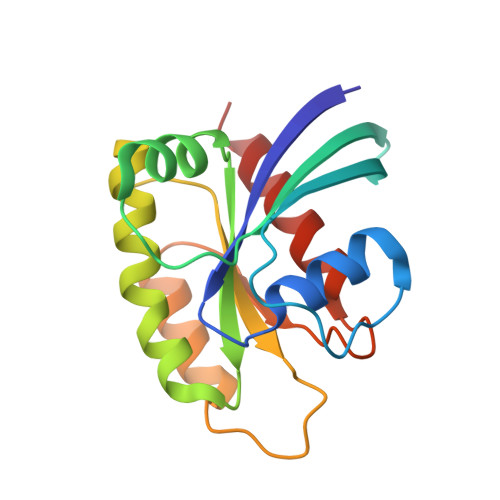
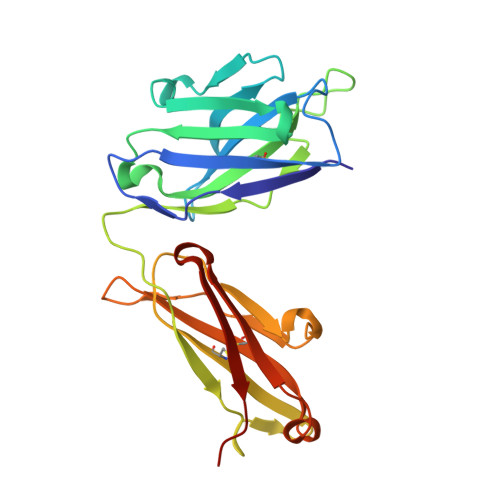
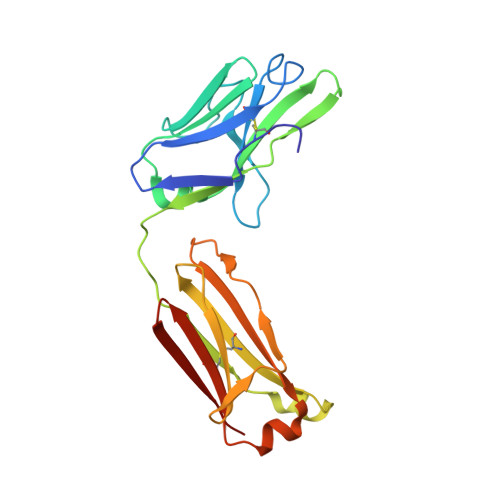
Conformation-locking antibodies for the discovery and characterization of KRAS inhibitors.
Davies, C.W., Oh, A.J., Mroue, R., Steffek, M., Bruning, J.M., Xiao, Y., Feng, S., Jayakar, S., Chan, E., Arumugam, V., Uribe, S.C., Drummond, J., Frommlet, A., Lu, C., Franke, Y., Merchant, M., Koeppen, H., Quinn, J.G., Malhotra, S., Do, S., Gazzard, L., Purkey, H.E., Rudolph, J., Mulvihill, M.M., Koerber, J.T., Wang, W., Evangelista, M.(2022) Nat Biotechnol 40: 769-778
- PubMed: 34992247
- DOI: https://doi.org/10.1038/s41587-021-01126-9
- Primary Citation of Related Structures:
7MDP, 7RP2, 7RP3, 7RP4 - PubMed Abstract:
Small molecules that stabilize inactive protein conformations are an underutilized strategy for drugging dynamic or otherwise intractable proteins. To facilitate the discovery and characterization of such inhibitors, we created a screening platform to identify conformation-locking antibodies for molecular probes (CLAMPs) that distinguish and induce rare protein conformational states. Applying the approach to KRAS, we discovered CLAMPs that recognize the open conformation of KRAS G12C stabilized by covalent inhibitors. One CLAMP enables the visualization of KRAS G12C covalent modification in vivo and can be used to investigate response heterogeneity to KRAS G12C inhibitors in patient tumors. A second CLAMP enhances the affinity of weak ligands binding to the KRAS G12C switch II region (SWII) by stabilizing a specific conformation of KRAS G12C , thereby enabling the discovery of such ligands that could serve as leads for the development of drugs in a high-throughput screen. We show that combining the complementary properties of antibodies and small molecules facilitates the study and drugging of dynamic proteins.
Organizational Affiliation:
Department of Antibody Engineering, Genentech, Inc., South San Francisco, CA, USA.