A Biparatopic Antibody-Drug Conjugate to Treat MET-Expressing Cancers, Including Those that Are Unresponsive to MET Pathway Blockade.
DaSilva, J.O., Yang, K., Surriga, O., Nittoli, T., Kunz, A., Franklin, M.C., Delfino, F.J., Mao, S., Zhao, F., Giurleo, J.T., Kelly, M.P., Makonnen, S., Hickey, C., Krueger, P., Foster, R., Chen, Z., Retter, M.W., Slim, R., Young, T.M., Olson, W.C., Thurston, G., Daly, C.(2021) Mol Cancer Ther 20: 1966-1976
- PubMed: 34315762
- DOI: https://doi.org/10.1158/1535-7163.MCT-21-0009
- Primary Citation of Related Structures:
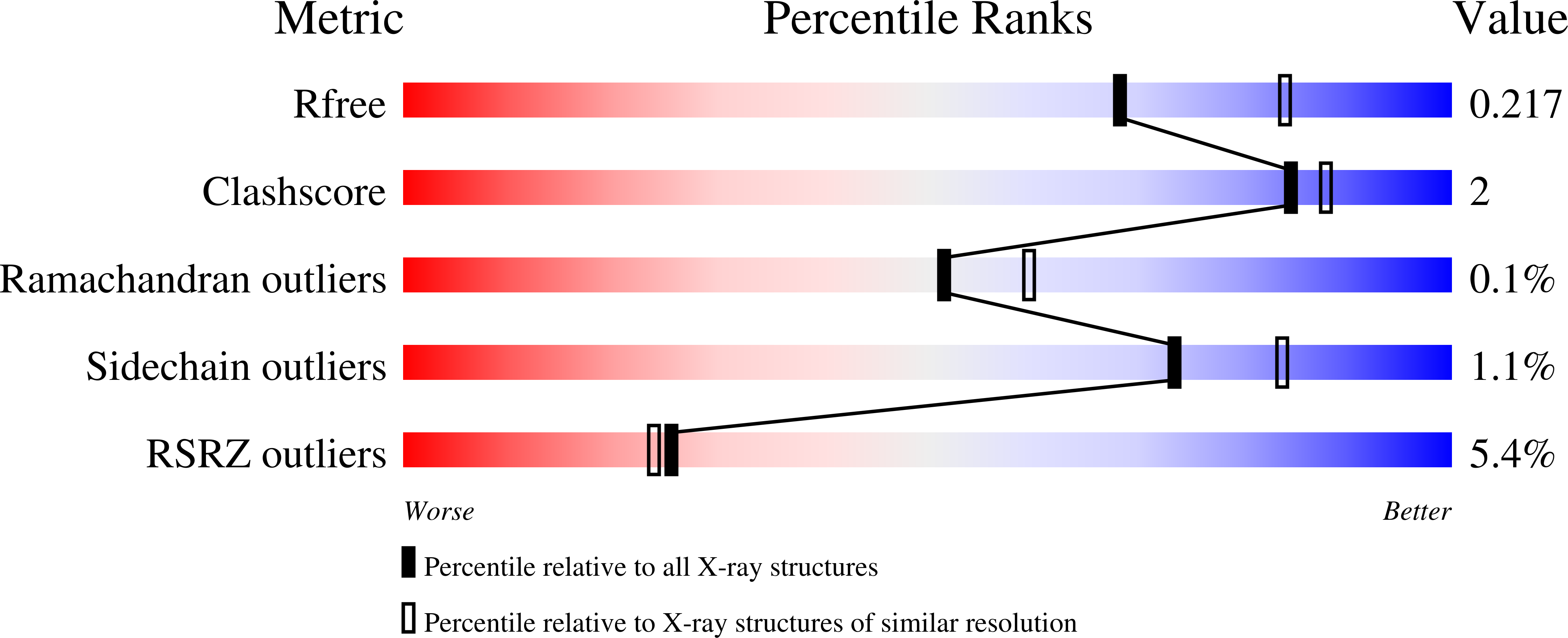
7L05 - PubMed Abstract:
Lung cancers harboring mesenchymal-to-epithelial transition factor ( MET ) genetic alterations, such as exon 14 skipping mutations or high-level gene amplification, respond well to MET-selective tyrosine kinase inhibitors (TKI). However, these agents benefit a relatively small group of patients (4%-5% of lung cancers), and acquired resistance limits response durability. An antibody-drug conjugate (ADC) targeting MET might enable effective treatment of MET-overexpressing tumors (approximately 25% of lung cancers) that do not respond to MET targeted therapies. Using a protease-cleavable linker, we conjugated a biparatopic METxMET antibody to a maytansinoid payload to generate a MET ADC (METxMET-M114). METxMET-M114 promotes substantial and durable tumor regression in xenografts with moderate to high MET expression, including models that exhibit innate or acquired resistance to MET blockers. Positron emission tomography (PET) studies show that tumor uptake of radiolabeled METxMET antibody correlates with MET expression levels and METxMET-M114 efficacy. In a cynomolgus monkey toxicology study, METxMET-M114 was well tolerated at a dose that provides circulating drug concentrations that are sufficient for maximal antitumor activity in mouse models. Our findings suggest that METxMET-M114, which takes advantage of the unique trafficking properties of our METxMET antibody, is a promising candidate for the treatment of MET-overexpressing tumors, with the potential to address some of the limitations faced by the MET function blockers currently in clinical use.
Organizational Affiliation:
Regeneron Pharmaceuticals, Inc., Tarrytown, New York. john.dasilva@regeneron.com.